Bridged nucleic acids (BNAs) are artificial bicyclic oligonucleotides that contain a five-membered or six-membered bridged structure with a “fixed” C3’-endo sugar puckering (Saenger 1984). The bridge is synthetically incorporated at the 2’, 4’-position of the ribose to afford a 2’, 4’-BNA monomer. The monomers can be incorporated into oligonucleotide polymeric structures using standard phosphoamidite chemistry.
BNAs are structurally rigid oligo-nucleotides with increased binding affinities and stability. Oligonucleotide modifications are characterized by the presence of one or more bicyclic ribose analogs. The structural similarity to native nucleic acids and the presence of a nitrogen atom within the bicyclic ring leads to very good solubility in water and allows for easy handling of synthetic primers and probes.
In contrast to peptide nucleic acids (PNAs) and minor groove binders (MGBs), but similar to LNAs BNA monomers can be used for both primers and probes in real time quantitative polymerase chain reaction (RT-Q-PCR) assays.
Compared to locked nucleic acids (LNAs) the substitution of DNA monomers with BNA monomers in oligonucleotides adds exceptional biological stability, resistance to nucleases and a significantly increased affinity to their complementary DNA targets.
The thermal stability is depending on the number of BNA monomers present in the sequence. Furthermore, BNA modifications greatly increase the melting temperature of oligonucleotides. Differences in melting temperatures (Tm) between perfectly and imperfectly matched nucleic acid duplexes allow for the discrimination even of single base mutations.
BNA-containing oligonucleotides useful for RT-Q-PCR assays can range in length between 12 and 20 nucleotides (nt), which is significantly shorter than unmodified primers and probes displaying the same Tm.
Furthermore, primers and probes modified with BNA monomers provide greater flexibility in designing consensus primers and probes for the detection of partially homologous target sequences, such as related viral species and serotypes. Finally, BNA-modified oligonucleotides in RT-Q-PCR assays reveal a specificity and sensitivity superior to other types of primers and probes.
Action mechanism of antisense oligonucleotides
The proposed action mechanism for antisense oligonucleotides may involve translation arrest, mRNA degradation mediated by RNase H and splicing arrest. This is illustrated in the following figure.
Design of BNANC [NH & NMe] Oligonucleotides
The placing of BNA NC [NMe] monomers in oligonucleotides, both, for DNA and RNA, is shown based on examples from published experimental data!
N = position of BNA NC [NMe] monomer.
Probes are usually labeled with Cy3 or FITC at the 5’ or 3’ end but other dyes may be used as well.
PCR Clamping using BNA oligonucleotides
More details on design and experimental results:
1. Gene silencing by targeting mRNA usingBNANC[NMe]’s !
Design of BNANC oligonucleotide: 5’-NNNnnnnnnnnNNN-3’,
where N depicts the position of a BNAnc monomer, and n depicts the position of a natural DNA. The use of BNANC’s increased nuclease resistance and improved overall properties for gene silencing by targeting mRNA. Buffer used: 100 mM NaCl, 10 mM sodium phosphate pH 7.2 for thermal melting studies. In vitro transfection was done with Lipofetamine 2000.
2′,4′-BNANC–based AONs are promising therapeutic agents for antisense therapy!! BNANC’s for gene silencing by mRNA targeting!
Yamamoto et al. 2012 showed that 2’,4’-BNANC-based AONs targeting apoB mRNA have higher binding affinities to the target RNA than do 2’,4’-BNA/LNA-based AONs. Additionally, in vitro transfection studies revealed the superior silencing effect of short 2’,4’-BNANC-based AONs (<20-ntlong), indicating that 2’,4’-BNANC may have advantageous properties as short antisense drugs. The BNANCs corresponding to the LNAs showed stronger inhibitory activities. Shorter AONs (13- to 14mers) showed better inhibitory activities. The 2’,4’-BNANC-14mer worked the best.
2. Gene silencing by targeting mRNA!
Design of BNANC oligonucleotide: 5′-NNnnnNNNnNnnnnnNnNNn-3′
Yamamoto et al. 2012: In vitro and in-vivo study. BNANC[NMe]’s with cholesterol lowering action targeting PCSK9 and with increased nuclease resistance and improved properties for gene silencing by targeting mRNA. Yamamoto and colleges achieved a dose-dependent decrease in serum LDL-C levels by using a 2′,4′-BNANC–based AON (P901SNC). Serum HDL-C levels and the levels of liver and kidney toxicity indicators were not elevated. Histopathological analysis revealed no severe hepatic toxicities. They also showed that a 2′,4′-BNANC–based AON (P901SNC) has greater potential to inhibit PCSK9 and to reduce serum cholesterol levels with no toxicity than a conventional 2′,4′-BNA–based AON. The high-potency and low-toxicity characteristics of a 2′,4′-BNANC–based AON were previously reported to effectively inhibit PTEN mRNA without elevation of the serum ALT level, whereas elevated serum ALT was observed in the 2′,4′-BNA counterpart-treated arm. Thus, it was concluded that 2′,4′-BNANC–based AONs can be a promising therapeutic agent for antisense therapy.
3. Interference RNA: => siRNA: BNANC’s for siRNA - RNAi => siRNA
Design of BNANC oligonucleotides that worked best:
5’-NNNnNnnnnnnnnnnnnnnnn-3’,
5’-nnNnnnNnnnNnnnNnnnNnn-3’
5’-nnnnnnnnnnNnNNnnnnnnn-3’,
5’-nnNnnNnnnnNnnnnNnnnnn-3’
Rahman and colleges in 2010 found that 2’,4’-BNA- and 2’,4’-BNANC-modified siRNAs are equally compatible with the RNAi machinery similar to that observed for natural siRNA. To improve siRNA biostability, a number of bridged nucleotide moieties can be incorporated in the sense strand without loss of the usual gene silencing property. Thermally stable functional siRNAs can also be obtained by slightly modifying the middle of the sense and antisense strands together. Unlike the 3’-overhang modification, this modification increased Tm satisfactorily and contains an antisense strand with BNA residues which might be more efficacious in gene silencing. Modification at the Ago2 cleavage site (9–11th positions) produced variable results based on siRNA composition and sequence; usually the modification at the 10th position of the sense strand is more sensitive. Modification at the 11th position of the cleavage site is safer than that of the 10th or 9th position. For the first time, this study as a whole shows the utility and capability of 2’,4’-BNANC, a highly stable and efficient nucleic acid derivative in RNAi technology, and also gives some new ideas about designing biostable, functional siRNAs consisting both of 2’,4’-BNA and 2’,4’-BNANC residues.
4. Antisense oligonucleotide targeting PTEN mRNA
Design of BNANC oligonucleotide:
5’-NNnnnnnnnnnnNN-3’ 14mer
43 d(CUTAGCACTGGCCU)30 20,40-BNANC[NMe] PTEN
Prakash et al in 2010.
5. Aptamer Capping at the 3’-end
A thrombin aptamer was capped with BNAs at the 3’-end. This increased nuclease resistance and the stability of the aptamer. Kasahara et al. 2010.
6. Synthesis of Novel 2’,4’-BNANC [NMe] nucleic acid analog.
Duplex formation: 5-d(GCGTTTTTTGCT)
Target ssRNA 5’-r(AGCAAAAAACGT)-3’, Tm = 63
Target ssdNA 5’-r(AGCAAAAAACGT)-3’, Tm = 51
Triplex formation: 5-d(TTTTTCTTTCTCT) 2’,4’-BNANC [NH] and LNA are better.
Target: dsDNA 5’-GCTAAAAAGAAAGAGAGATCG)-3’
3’-CGATTTTTCTTTCTCTCTCTAGC)-5’
Rahman et al. 2008.
A list of rules for the design of probes using BNAs
| BNAs |
| (i) |
Place one BNA monomer per every 3 bases within an oligomer.
For example in a 20mer oligomer, about 4 BNA monomers can be inserted (there is some degree of freedom as to the exact positions, in other words, they do not have to be exactly every 3 to 6 bases).
According to the specific application, the mode of BNA modification may need to be changed. Both gapmer- and chimera-modification with BNA-NC will be effective for antisense application. More than four continuous natural DNA monomers as part in a BNA-modified oligonucleotide was found to be necessary to recruit RNase H. For diagnostic application, the modification of one bp with a BNA nucleotide maybe enough in some cases.
|
| (ii) |
The spacing of the BNA monomers need not be different within the oligo for different applications. For example, if the spacings are appropriate the same monomer may be used. |
| (iii) |
The use of no more than 4-8 BNA’s within a 20 mer probe is recommended but is depended on the specific application. |
| (iv) |
Each BNA –NC monomer increase the Tm by about 4 degrees Celsius. Therefore this can be used to estimate the Tm of the BNA containing oligonucleotide). I agree. |
| (v) |
An oligonucleotide designed with 2’, 4’-BNA-NC[NH] is more selective to ssRNA and binds more strongly than LNA if more bases are modified. |
| (vi) |
An oligonucleotide designed with 2’, 4’-BNA-NC[NH] binds also to ssDNA and binds also more strongly than LNA if more bases are modified. The ssDNA binding strength of BNA-NC(NH) is equivalent to (or slightly more than) that of LNA. |
| (vii) |
BNA-NC(NMe) modification add a very high nuclease-resistance to the oligonucleotide (much more than LNA modification). This property is desirable not only for therapeutic application but also for diagnostic use. |
|
References
Kazuyuki Miyashita, S. M. Abdur Rahman, Sayori Seki, Satoshi Obikaab and Takeshi Imanishi; N-Methyl substituted 2’,4’-BNANC: a highly nuclease-resistant nucleic acid analogue with high-affinity RNA selective hybridization. Chem. Commun., 2007, 3765–3767.
S. M. Abdur Rahman, Sayori Seki, Satoshi Obika, Sunao Haitani, Kazuyuki Miyashita, and Takeshi Imanishi; Highly Stable Pyrimidine-Motif Triplex Formation at Physiological pH Values by a Bridged Nucleic Acid Analogue. Angew. Chem. Int. Ed. 2007, 46, 4306–4309.
S. M. Abdur Rahman, Sayori Seki, Satoshi Obika, Haruhisa Yoshikawa, Kazuyuki Miyashita, and Takeshi Imanishi; Design, Synthesis, and Properties of 2‘,4‘-BNANC: A Bridged Nucleic Acid Analogue. J. AM. CHEM. SOC. 2008, 130, 4886-4896.
S. M. Abdur Rahman, Hiroyuki Sato, Naoto Tsuda, Sunao Haitani, Keisuke Narukawa, Takeshi Imanishi, Satoshi Obika; RNA interference with 2’,4’-bridged nucleic acid analogues. Bioorganic & Medicinal Chemistry 18 (2010) 3474–3480.
Saenger, W.; Principles of Nucleic Acid Structure, Springer-Verlag, New York, 1984.
Torigoe H, Rahman SM, Takuma H, Sato N, Imanishi T, Obika S, Sasaki K.; 2'-O,4'-C-aminomethylene-bridged nucleic acid modification with enhancement of nuclease resistance promotes pyrimidine motif triplex nucleic acid formation at physiological pH.Chemistry. 2011 Feb 25;17(9):2742-51. doi: 10.1002/chem.201002745. Epub 2011 Jan 24.
Hidetaka Torigoe and Takeshi Imanishi; Chemical Modification of Oligonucleotides: A Novel Approach Towards Gene Targeting. Chapter 6 in Biochemistry, Genetics and Molecular Biology » "Mutagenesis", book edited by Rajnikant Mishra, ISBN 978-953-51-0707-1, Published: August 17, 2012 under CC BY 3.0 license. DOI: 10.5772/50393
Tsuyoshi Yamamoto, Hidenori Yasuhara, FumitoWada, Mariko Harada-Shiba, Takeshi Imanishi, and Satoshi Obika; Superior Silencing by 2’,4’-BNANC-Based Short Antisense Oligonucleotides Compared to 2’,4’-BNA/LNA-Based Apolipoprotein B Antisense Inhibitors. Hindawi Publishing Corporation, Journal of Nucleic Acids Volume 2012, Article ID 707323, 7 pages. doi:10.1155/2012/
Tsuyoshi Yamamoto, Mariko Harada-Shiba, Moeka Nakatani, Shunsuke Wada, Hidenori Yasuhara, Keisuke Narukawa, Kiyomi Sasaki, Masa-Aki Shibata, Hidetaka Torigoe, Tetsuji Yamaoka, Takeshi Imanishi and Satoshi Obika; Cholesterol-lowering Action of BNA-based Antisense Oligonucleotides Targeting PCSK9 in Atherogenic Diet-induced Hypercholesterolemic Mice Molecular Therapy–Nucleic Acids (2012) 1, e22; oi:10.1038/mtna.2012.16.
www.biosyn.com/faq/bna-general-design-guidelines.aspx
http://www.biosyn.com/tew/bridged-nucleic-acid-bna3-design-guidelines.aspx
Design of allele specific primer
ABO genotyping:
Yaku, H., Yukimasa, T., Nakano, S.-i., Sugimoto, N. and Oka, H. (2008), Design of allele-specific primers and detection of the human ABO genotyping to avoid the pseudopositive problem. ELECTROPHORESIS, 29: 4130–4140. doi: 10.1002/elps.200800097
Mutation detection:
Morandi L, de Biase D, Visani M, Cesari V, De Maglio G, et al. (2012) Allele Specific Locked Nucleic Acid Quantitative PCR (ASLNAqPCR): An Accurate and Cost-Effective Assay to Diagnose and Quantify KRAS and BRAF Mutation. PLoS ONE 7(4): e36084. doi:10.1371/journal.pone.0036084.
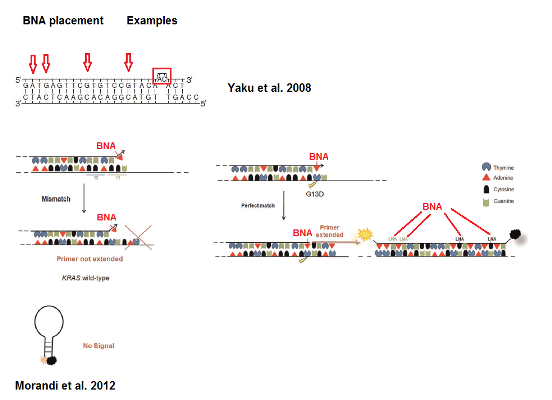
Figure 1: Examples for placements of BNA for allele specific probes. The red arrows, box and lines indicate the recommended placement of the BNA monomers. Modified after Morandi et al. 2012.
Note: Design may vary and will have to be experimentally tested.
In general BNA’s can be placed similar to LNA’s. However, to increase the binding affinity (= Increased Melting Temperature) more BNA monomers can be added. Adding BNA’s at G’s and T’s are the best positions for this.
To make the probes more specific for allele detection one or two mismatches can be incorporated at the 3’ end.
Furthermore, adding S atoms at the phosphor moieties at positions 1, 2, and 3 downstream from the 5’-end may further improve the stability and nuclease resistance of the probe.
|