In cells, RNA transcripts are bound to proteins to form RNA binding protein complexes (RBPs). RBPs play essential roles in regulating cellular processes, such as RNA metabolism, trafficking, mRNA splicing, localization, translation, and degradation. The interaction of coding and non-coding RNAs with RNA-binding proteins regulate critical cellular processes post-transcriptionally. The 3’-untranslated regions (3'-UTRs) contain regulatory motifs for RNA-binding protein involved in the control of mRNA expression levels and stability.
In recent years, scientists developed many strategies to analyze complex and dynamic RNA-protein networks in cells. However, identifying proteins and other molecules that specifically bind to RNAs for studying their biological function is challenging.
Specific RNA affinity probes allow the study of RNA interactions with molecules such as DNA, RNA, and proteins. For example, synthetic probes allow localization of a gene or a DNA and RNA sequence within a biological context. Historically, for this approach, a small piece of DNA or RNA labeled with a radioisotope (3H, 32P, or 125I), a fluorescent dye, or a ‘tag’ that can be recognized by a specific antibody, has been used for localization of the particular target, a gene sequence, via hybridization. These probes are known as hybridization probes. Biotin is now a common tag used for many applications.
" Bio-Synthesis provides a full spectrum of high quality custom oligonucleotide modification services including biotinylation by direct solid-phase chemical synthesis or enzyme-assisted approaches to obtain artificially modified oligonucleotides, such as mRNAs or siRNAs, containing natural or modified backbone, base, sugar and internucleotide linkages. Bio-Syntheis also provides biotinylated mRNA".
Synthetic biotinylated miRNA probes
Biotin tagged synthetic microRNA (miRNA) duplexes allow the identification of messenger RNAs (mRNAs) associated with miRNAs.
Ørom and Lund, in 2007, showed that the production of synthetic miRNA duplexes enables the identification of mRNAs associated with miRNAs. The approach involved producing synthetic miRNA duplexes carrying a biotin group at the 3’-end of the miRNA sense strand. The tagged miRNA sense strand incorporates into the miRISC complex and associates with endogenous target mRNAs after cell transfection. Mild cell lysis allows the capture of the miRNA-mRNA complexes on streptavidin beads now ready for purification and analysis.
Biotin used in combination with the proteins avidin or streptavidin also enables labeling gene segments. These “pull-down probes” allow the purification of molecular complexes associated with the targeted DNA or RNA sequence. Reactive derivatives of biotins such as para-nitro-phenyl or N-hydroxy-succinimide esters enable the chemical covalent attachment or conjugation of biotin to side-chain groups of proteins, polysaccharides, oligonucleotides, as well as others. A variety of biotinylation reagents are now commonly available from various companies.
Enzymatic biotinylated RNA tags
RNA transcripts containing a short, encoded hairpin recognition motif can be enzymatically labeled.
Recently, Busby et al. showed that RNA transcripts containing a short, encoded hairpin recognition motif could be enzymatically labeled. The labeling reaction utilizes a preQ1-biotin tag and bacterial tRNA guanine transglycosylase (TGTase). The natural derivative preQ1 allows marking the RNA sequence of interest containing a TGT recognition hairpin. PreQ1 is a metabolic intermediate in the synthetic pathway that produces the hypermodified guanine nucleotide, queuosine (Q). In eubacteria, TGTase inserts preQ1 into the wobble position of specific tRNAs, modified further to give queuosine.
Biotinylation RNA can be efficiently purified using avidin or streptavidin beads. Bacterial TGT covalently labels RNAs containing a short, encoded hairpin recognition motif using nucleobase derivatives.
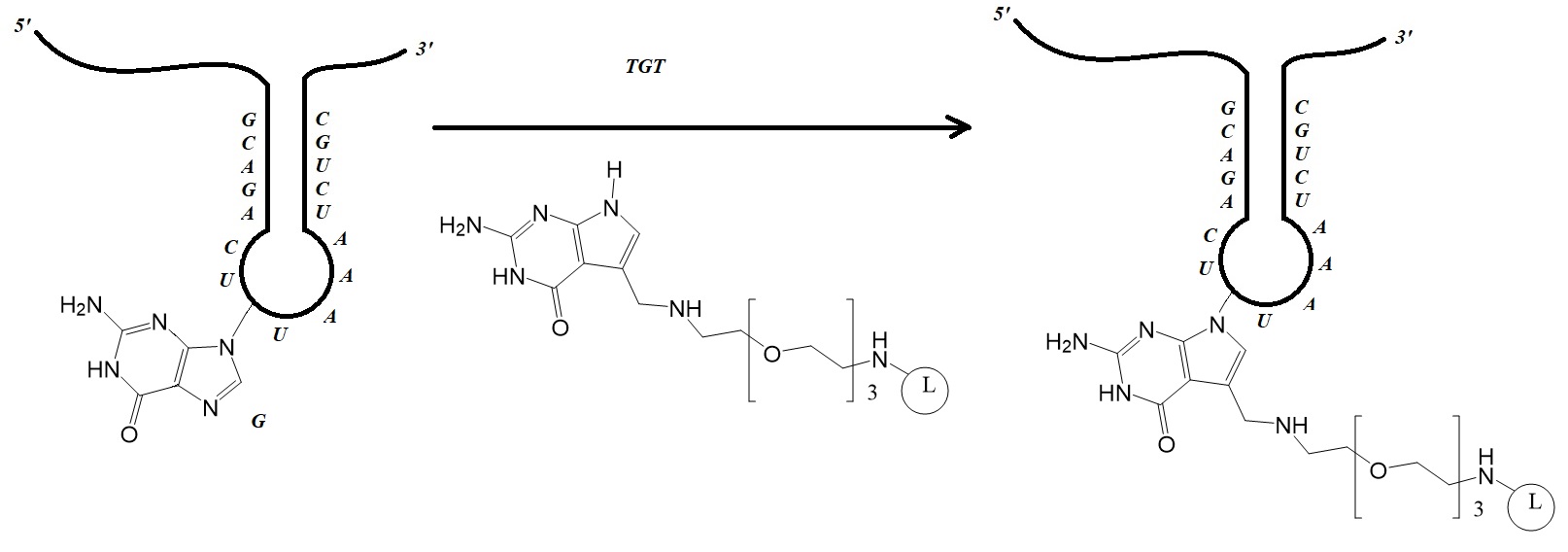
Figure 1: Site-specific incorporation of nucleobase derivatives. Derivatives can be affinity labels or fluorophores (L), including biotin, BODIPY, thiazole orange, and Cy7 linked through a PEG linker attached to PreQ1 (Adapted from Alexander et al.).
tRNA-guanine transglycosylase (TGT, EC 2.4.2.29) catalyzes a base-exchange reaction resulting in an anticodon modification of certain tRNAs. Prokaryotic TGT replaces guanine (G) with 7-aminomethyl-7-deazaguanine (PreQ1) at the wobble position of four specific tRNAs.
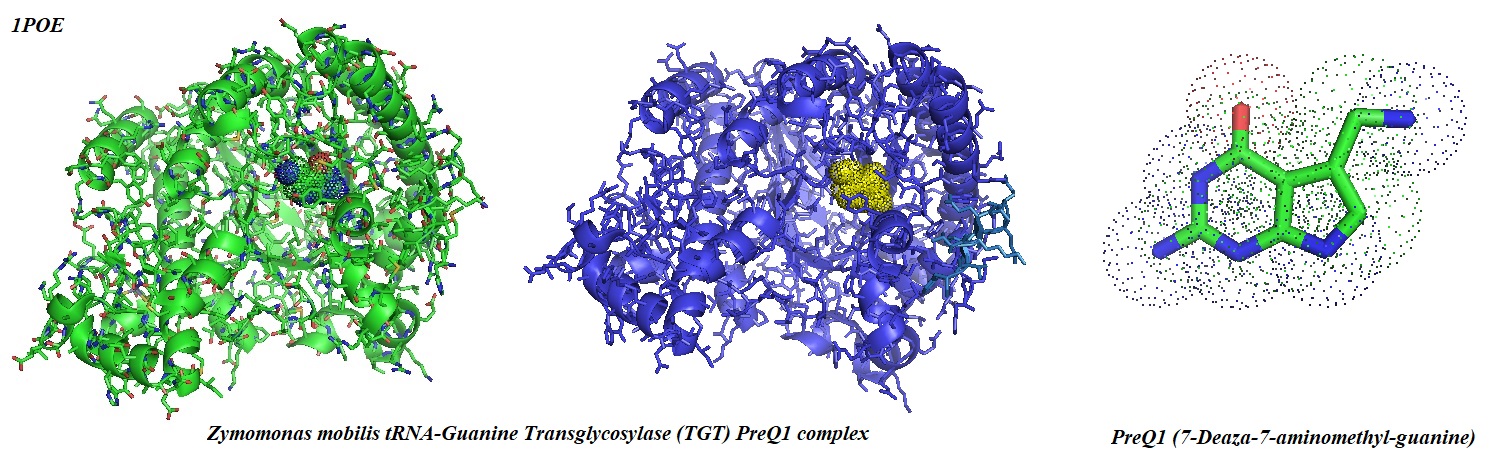
Figure 2: Crystal structure of the Zymomonas mobilis tRNA-guanine transglycosylase in complex with PreQ1 (Brenk et al.). The substrate PreQ1 is shown in the binding pocket as a doted structure (left) and as a yellow dot structure (middle) in the binding pocket of the enzyme.
Queuosine is a nucleoside found in tRNA with an additional cyclopentenyl ring added via an amino group to the methyl group of 7-methyl-7-deazaguanosine. This nucleoside is present in the first position of the anticodon of tRNA-tyrosine, tRNA-histidine, tRNA-asparagine, and tRNA-aspartic acid found in many organisms. Queuosine may play a role in the regulatory function of tRNA.
The RNA methyltransferase Dnmt2‐dependent tRNA methylation is dynamically modulating queuosine‐tRNA levels. Queuosine‐tRNA levels control the translation speed of queuosinylated‐tRNA decoded codons. An altered translation in the absence of queuosine results in misfolded aggregates. These aggregates trigger endoplasmic reticulum stress and the unfolded protein response.
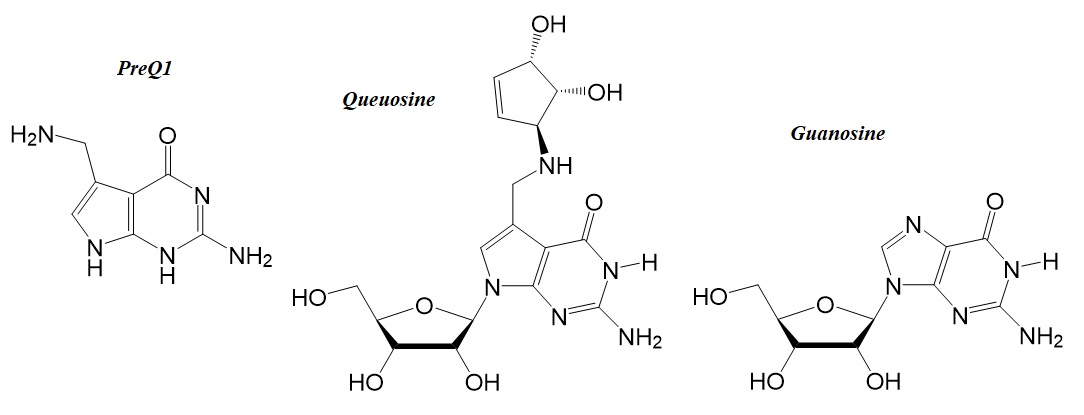
Figure 3: Chemical structures of PreQ1, queuosine (Q), and guanosine (G).
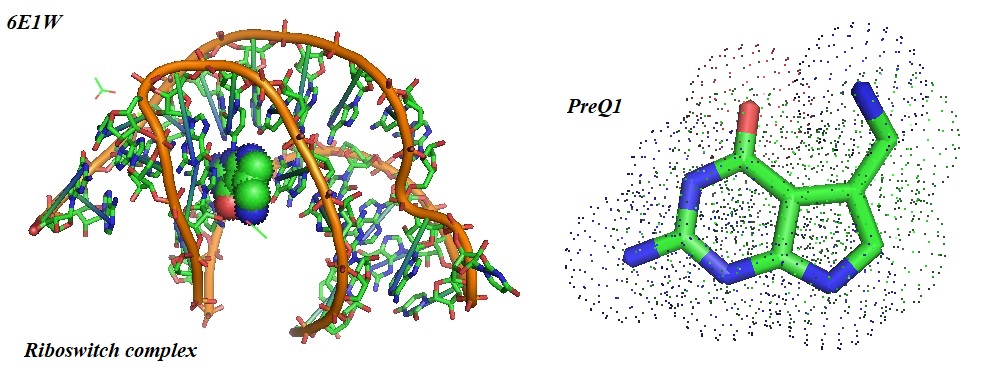
Figure 4: Crystal structure of a class I PreQ1 riboswitch in complex with PreQ1 (left) and the structural model or PreQ1 (right). Riboswitches are natural RNA aptamers regulating gene expression by binding to specific small molecules. Connelly et al., in 2019, reported the discovery of synthetic small molecules that specifically bind to the PreQ1 riboswitch aptamer. Alteration of the chemical structure of the ligand was found to cause changes in the mode of RNA binding affecting regulatory functions.
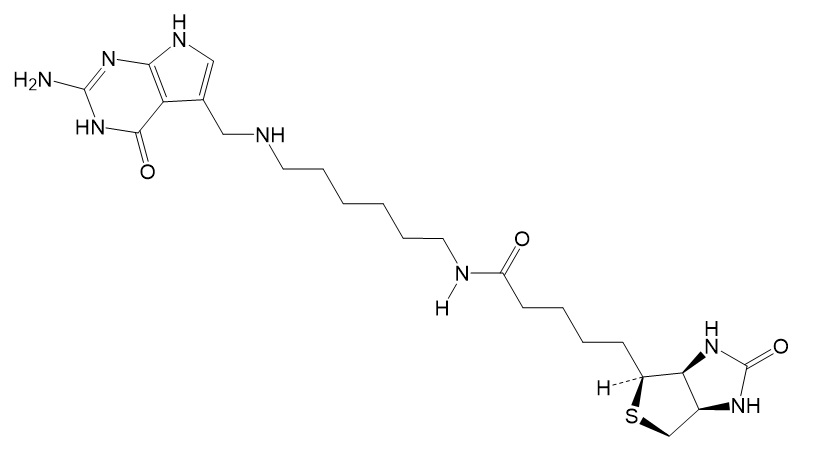
FIGURE 5: PreQ1-biotin. RNA molecules containing a TGT recognition hairpin can be labeled with the preQ1-biotin deivative using bacterial tRNA Guanine Transglycosylae (TGT).
Xie et al., in 2003, achieved trapping of a covalent intermediate of Zymomonas mobilis TGT and an RNA substrate by adding 9-deazaguanine (9dzG) to the reaction mixture. The purification and crystallization of the complex allow the research group to solve the intermediate X-ray structure (figure 6).
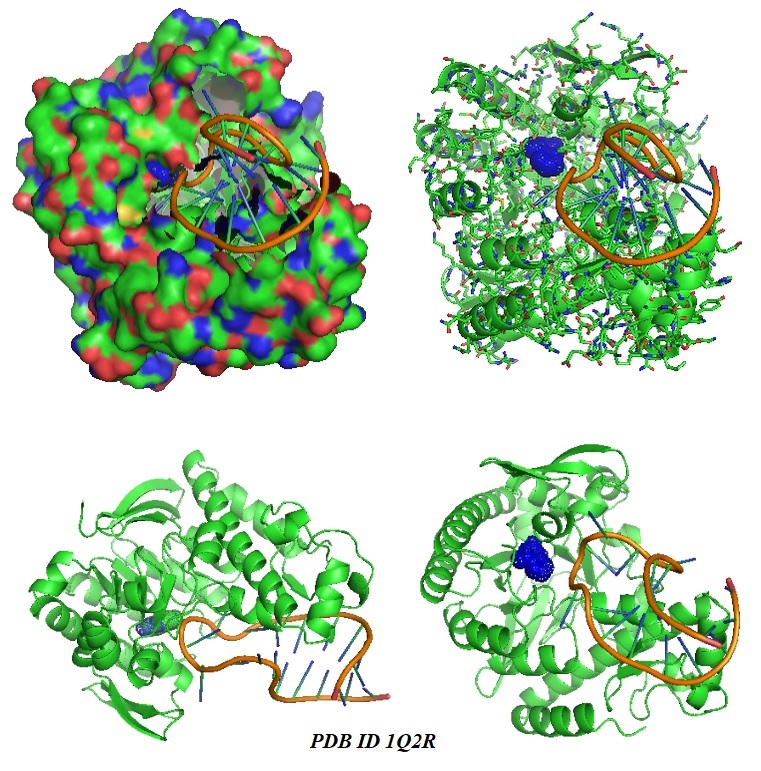
FIGURE 7: Crystal structure of the chemically trapped catalytic tRNA guanine transglycosylase covalent intermediate (Xie et al., 2003; PDB ID 1Q2R). Different models of the complex are shown. The structure shows the deglycosylated state of the guanine. 9-Deazaguanine is shown in blue.
Cap-biotinylated RNAs
Cap-biotinylated RNAs allow quantifying cap-dependent translation of mRNAs, mRNA processing, and turnover.
Co-transcriptional incorporation of cap analogs allows studying translation efficiency, capping efficiency, and the susceptibility to decapping by cap-specific mRNA-decapping pyrophosphatase, Dcp1–Dcp2. Furthermore, biotinylated caps can serve as capture probes. Bednarek et al., in 2018, reviewed a general synthesis strategy for the synthesis of biotinylated 5’-cap RNA analogs.
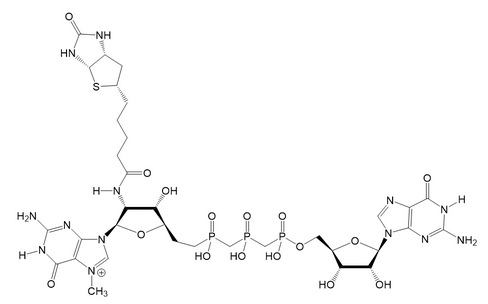
FIGURE 7: Structure of a biotinylated cap (Adopted from Bednarek et al.). Chemical synthesis allows the production of cap analogs with a modified triphosphate bridge as well. The biotin affinity tag is attached to the 2’-position of m7G via an amide bond.
According to Nierves & Lange, biotin labels or tags allow detection sites of biotinylation combined with mass spectrometry. This approach is beneficial for the proteomic study of biotinylated peptides. Enzymatic site-specific labeling of RNA targets is possible with RNA guanine transglycosylase from E. coli using unnatural nucleobase substrates. Busby et al. recently demonstrated the identification of RNA–protein interactions and the selective enrichment of cellular RNA in mammalian systems. The research group showed that an engineered enzyme variant achieved high labeling selectivity levels against the human transcriptome. The research group observed a 145-fold enrichment of cellular RNA directly isolated from mammalian cell lysates.
Reference
Alexander, S. C.; Busby, K. N.; Cole, C. M.; Zhou, C. Y.; Devaraj, N. K. Site-Specific Covalent Labeling of RNA by Enzymatic Transglycosylation. J. Am. Chem. Soc. 2015, 137 (40), 12756–12759. [PubMed]
Sylwia Bednarek, Vanesa Madan, Pawel J. Sikorski, Ralf Bartenschlager, Joanna Kowalska and Jacek Jemielity; mRNAs biotinylated within the 5′ cap and protected against decapping: new tools to capture RNA–protein complexes. Philosophical Transactions of the Royal Society B: Biological SciencesVolume 373, Issue 1762. [The Royal Socieity]
Bioconjugation for drug discovery
Ruth Brenk, Milton T Stubbs, Andreas Heine, Klaus Reuter, Gerhard Klebe; Flexible adaptations in the structure of the tRNA-modifying enzyme tRNA-guanine transglycosylase and their implications for substrate selectivity, reaction mechanism and structure-based drug design. Chembiochem. 2003 Oct 6;4(10):1066-77. [PubMed]
Kayla N. Busby, Amitkumar Fulzele, Dongyang Zhang, Eric J. Bennett, and Neal K. Devaraj; Enzymatic RNA biotinylation for affinity purification and identification of RNA-protein interactions. ACS Chemical Biology 2020 15 (8), 2247-2258 . [bioRxiv]
Connelly CM, Numata T, Boer RE, Moon MH, Sinniah RS, Barchi JJ, Ferré-D'Amaré AR, Schneekloth JS Jr. Synthetic ligands for PreQ1 riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure. Nat Commun. 2019 Apr 2;10(1):1501. [PMC]
Jenkins JL, Krucinska J, McCarty RM, Bandarian V, Wedekind JE. Comparison of a preQ1 riboswitch aptamer in metabolite-bound and free states with implications for gene regulation. J Biol Chem. 2011 Jul 15;286(28):24626-37. [PMC]
D. Metzler. Biochemistry. The chemical reactions of living cells. 2nd Edition). [Book]
Lorenz Nierves and Philipp F. Lange; DETECTABILITY OF BIOTIN TAGS BY LC-MS/MS. [bioRxiv]
Ulf Andersson Ørom and Anders H. Lund; Isolation of microRNA targets using biotinylated synthetic microRNAs. Methods 43, 2, 162-165. [PubMed] [Europe PMC]
Queuosine
Francesca Tuorto, Carine Legrand, Cansu Cirzi, Giuseppina Federico, Reinhard Liebers, Martin Müller, Ann E Ehrenhofer‐Murray, Gunnar Dittmar, Hermann‐Josef Gröne, Frank Lyko; Queuosine‐modified tRNAs confer nutritional control of protein translation. EMBO J (2018)37:e99777 [EMBO]
Winkler W. C., Breaker R. R.; Regulation of bacterial gene expression by riboswitches. (2005) Annu. Rev. Microbiol. 59, 487–517. [PubMed]
Xie W, Liu X, Huang RH. Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate. Nat Struct Biol. 2003 Oct;10(10):781-8. doi: 10.1038/nsb976. Epub 2003 Aug 31. Erratum in: Nat Struct Biol. 2004 Jul;11(7):678. [PubMed]
---...---
Bio-Synthesis provides a full spectrum of high quality custom oligonucleotide modification services including biotinylation by direct solid-phase chemical synthesis or enzyme-assisted approaches to obtain artificially modified oligonucleotides, such as mRNAs or siRNAs, containing natural or modified backbone, base, sugar and internucleotide linkages.
Bio-Synthesis specializes in complex oligonucleotide modifications using phosphodiester backbone, purine and pyrimidine heterocyclic bases, and sugar modified nucleotides such as our patented 3rd generation Bridged Nucleic Acids.
Furthermore, Bio-Synthesis specializes in biomolecular conjugation for drug discovery.
.jpg)
---...---