Ribose 2’-O-methylation is a molecular signature of self and non-self.
Messenger RNAs (mRNAs) of higher eukaryotes have 5’-cap structures with ribose 2’-O-methylation. The mRNA can occur methylated at the 2’-O position of the first and second nucleotide upstream of the 5’-cap, known as cap 1 and cap 2. Uncapped RNA molecules, for example, nascent viral transcripts, may be detected as "non-self" by host cells. This event triggers an antiviral innate immune response by producing interferons. In addition, the innate immune system also discriminates between self and non-self via the detection of inosine sites in the epitranscriptome with the help of ADAR proteins.
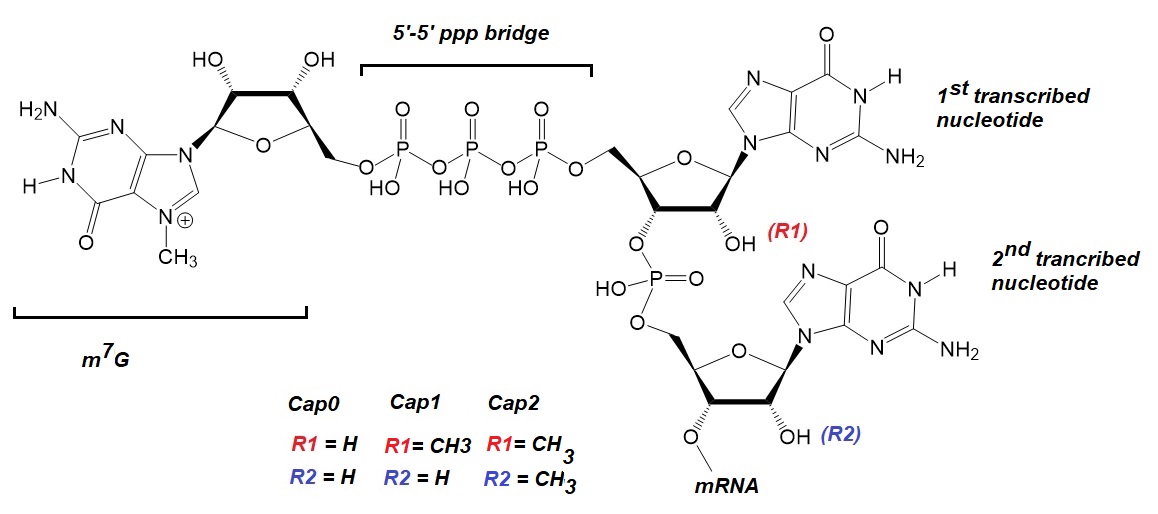
Figure 1: RNA cap structure: The Cap 0 structure consists of a guanosine residue, methylated in the N-7 position bound to the terminal 5’-end nucleotide of the mRNA via a 5’-5' triphosphate bridge. Subsequent 2’-O-methylation in the ribose of the first, or both the first and the second, transcribed mRNA nucleotides generates cap1 or cap2, respectively. Adapted from Belenage et al.
{ Bélanger, F.; Stepinski, J.; Darzynkiewicz, E.; Pelletier, J. Characterization of hMTr1, a human Cap1 2’-O-ribose methyltransferase. J. Biol. Chem. 2010, 285, 33037–33044 [PMC]; Dilyana G. Dimitrova, Laure Teysset and Clément Carré; RNA 2’-O-Methylation (Nm) Modification in Human Diseases. Genes 2019, 10, 117 [PMC] }
Züst et al., in 2011, demonstrated that human and mouse coronavirus mutants that do not have a 2’-O-methyltransferase induced a higher type I interferon expression and were sensitive to type I interferon. The study showed that the 2′-O-methylation of mRNA provides a molecular signature with a dual role in the interaction with host innate immune responses.
2’-O-Methylation of mRNA protects viral RNA from recognition by melanoma differentiation-associated protein 5 (Mda5). Mda5 functions as a pattern recognition receptor and detects double-stranded RNA (dsRNA). The presence of the modification prevents Mda5-dependent production of type I interferon in virus-infected cells.
2’-O-Methylation of viral mRNA allows the virus to evade interferon-mediated restriction of viral replication. 2’-O-methylation of viral mRNA is vital in innate immune responses in host cells. The methylation status in DNA allows distinction between self and non-self-nucleic acids since methylated CpG dinucleotide motifs in DNA activate Toll-like receptor 9. When Mda5 detects the absence of 2’-O-methylation in the mRNA, it initiates an immune response. A functioning innate immune response in the host relies on the reliable detection of pathogens. This immune response allows the host to limit pathogen replication and spread.
The researcher studied coronaviruses because coronaviruses encode their own 5′ mRNA cap-methylation machinery. This feature allows the investigation of recombinant viruses' phenotype with mutant 2′-O-methyltransferase proteins. The study showed that 2′-O-methyltransferase activity is associated with the viral nonstructural protein nsp16. The nsp16 protein is highly conserved among coronaviruses and an integral subunit of the viral replicase-transcriptase complexes located at virus-induced double-membraned vesicles in the host cell cytoplasm.
Züst et al. produced a mutant by substituting alanine for the aspartic acid at position 129 of the highly conserved catalytic KDKE tetrad of nsp16 (HCoV-D129A). The substitution result was a completely deactivated 2′-O-methyltransferase activity in recombinant, bacteria-expressed nsp16 proteins of feline coronavirus and severe acute respiratory syndrome coronavirus. However, using the vaccinia virus 2′-O-methyltransferase VP39 in vitro again methylated poly(A)-containing RNA from HCoV-D129A-infected cells.
{ Züst R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, et al. (February 2011). "Ribose 2'-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5". Nature Immunology. 12 (2): 137–43. doi:10.1038/ni.1979. PMC 3182538. PMID 21217758. [Nature] }
Bio-Synthesis provides a full spectrum of high quality custom oligonucleotide modification services by direct solid-phase chemical synthesis or enzyme-assisted approaches to obtain artificially modified oligonucleotides containing backbone, base, sugar and internucleotide linkages including messenger RNA.
Bio-Synthesis specialize in complex oligonucleotide modifications using phosphodiester backbone, purine and pyrimidine heterocyclic bases, and sugar modified nucleotides such as our patented 3rd generation Bridged Nucleic Acids.
---...---