The new SARS coronavirus, which is the cause of the COVID-19 pandemic, hijacks the translation machinery in human host cells by converting Cap-0 to Cap-1 in the host's mRNA to avoid an innate immune response.
Intending to develop effective therapies for COVID-19, Viswanathan et al. studied the mechanisms permitting the virus to invade cells and evade the host's innate immune system. Viswanathan et al. in 2020 reported that the non-structural protein 16 (nsp16) methylates the 5’-end of virally encoded mRNAs to mimic cellular mRNAs. This modification hides the virus's mRNAs from the innate immune system. The research team solved the high-resolution structure of a ternary complex of full-length nsp16 and nsp10 of SARS-CoV-2 complexed to the cognate RNA substrate and a methyl donor, S-adenosyl methionine. The nsp16/nsp10 heterodimer's structural model revealed the methylation of the 2’-oxygen of the ribose sugar of the first nucleotide of SARS-CoV-2 mRNAs. Also, the structure revealed fundamental conformational changes in the RNA substrate binding site and the biochemistry of RNA Cap methylation. Furthermore, Viswanathan et al. identified a potential allosteric site as a target for developing antiviral therapies to treat SARS-CoV-2 infections.
The SARS-CoV-2 Nsp16 protein forms a complex with nsp10 to convert mRNA species from the Cap-0 (me7GopppA1) to the Cap-1 form (me7GopppA1m). The protein complex methylates the ribose 2’-OH group of the nascent mRNA's first nucleotide using S-adenosyl methionine (SAM) as the methyl donor molecule. In CoVs, the first nucleotide is usually adenosine. Cap-1 avoids induction of the innate immune response.
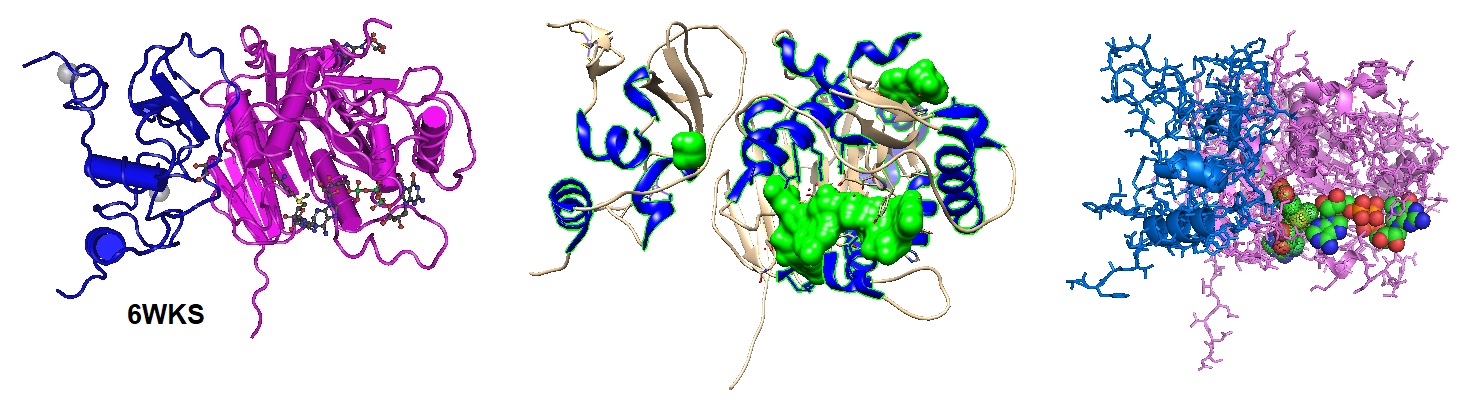
Figure 1: Different images of the SARS-CoV-2 nsp16/nsp10 in complex with RNA cap analog (m7GpppA) and S-adenosylmethionine (6WKS).
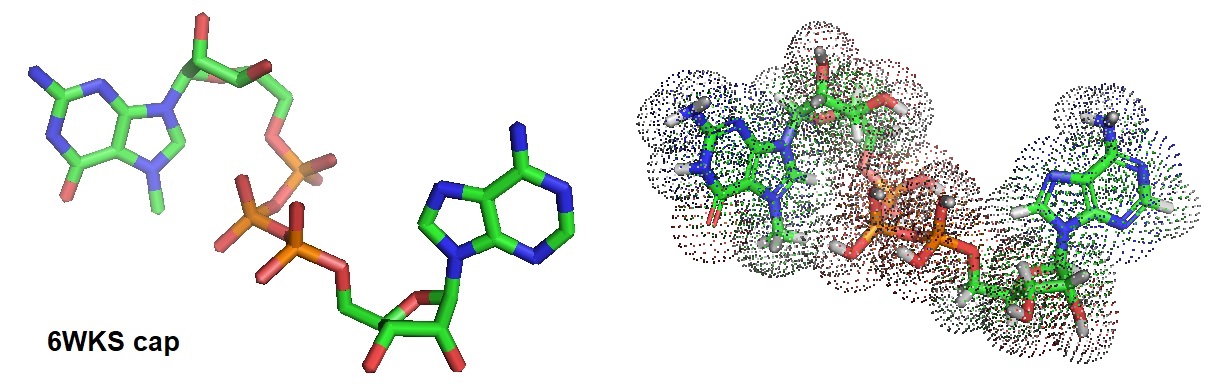
Figure 2: Configuration of m7GpppA as observed in the complex 6WKS.
Also, in 2020, Rosas-Lemus et al. determined the structures for nsp16-nsp10 heterodimers in complew with the methyl donor S-adenosylmethionine (SAM), the reaction product S-adenosylhomocysteine (SAH), or the SAH analog sinefungin (SFG). The research group solved structures for nsp16-nsp10 in complex with the methylated Cap-0 analog m7GpppA and either SAM or SAH. Comparing the structures with published structures for nsp16 from other beta coronaviruses revealed flexible loops in open and closed conformations at the m7GpppA-binding pocket.
Bound sulfates observed in several of the structures suggested the location of the ribonucleic acid backbone phosphates in the ribonucleotide-binding groove. The researchers also identified additional nucleotide-binding sites on the face of the protein opposite the active site.
Figure : Structures of nsp10-nsp16-SAM complex (6W4H).
Vaccinating mice with nsp16-defective SARS-CoV-1 or an immunogenic disruption of the nsp16-nsp10 interface protects mice from lethal infections. As a result, Viswanathan et al. suggested nsp16/nsp10 as an attractive drug target to treat COVID-19 patients.
Reference
Viswanathan T, Arya S, Chan SH, Qi S, Dai N, Hromas RA, Park JG, Oladunni F, Martinez-Sobrido L, Gupta YK. Structural Basis of RNA Cap Modification by SARS-CoV-2 Coronavirus. bioRxiv [Preprint]. 2020 Apr 26:2020.04.26.061705. [PMC] [6WKS]
Rosas-Lemus M, Minasov G, Shuvalova L, Inniss NL, Kiryukhina O, Brunzelle J, Satchell KJF. High-resolution structures of the SARS-CoV-2 2'-O-methyltransferase reveal strategies for structure-based inhibitor design. Sci Signal. 2020 Sep 29;13(651):eabe1202. [PubMed] [6W4H]
Teodoro Bottiglieri, S-Adenosyl-L-methionine (SAMe): from the bench to the bedside—molecular basis of a pleiotrophic molecule, The American Journal of Clinical Nutrition, Volume 76, Issue 5, November 2002, Pages 1151S–1157S. [pdf]
---...---
" Bio-Synthesis provides a full spectrum of high quality custom oligonucleotide modification services including back-bone modifications, conjugation to fatty acids, biotinylation by direct solid-phase chemical synthesis or enzyme-assisted approaches to obtain artificially modified oligonucleotides, such as BNA antisense oligonucleotides, mRNAs or siRNAs, containing a natural or modified backbone, as well as base, sugar and internucleotide linkages.
Bio-Synthesis also provides biotinylated mRNA and long circular oligonucleotides".
---...---