Nucleoside phosphoramidite monomers containing two protected hydroxyl functional groups utilized as building blocks allow the synthesis of branched oligonucleotides. During the biosynthesis of eukaryotic messenger RNA, branched oligoribonucleotides are naturally found as 'lariat structures' in splicing intermediates.
Branched DNA (bDNA) allows the design and synthesis of signal amplification assays to detect nucleic acid sequences in hybridization assays quantitatively. Examples are assays for the detection of early viral infection states in plasma. Sensitive assays are needed to measure small amounts of viral RNA as this is the case for Human Immunodeficiency Virus Type 1 (HIV-1) or SARS-CoV and SARS-CoV-2 (COVID-19) infections.
In addition, the incorporation of branched nucleotide monomers into DNA or RNA also allows the construction of complex DNA and RNA structures such as DNA origami or complex nanostructures based on nucleic acid sequences. Well-positioned branching monomers enable the synthesis of well-defined DNA or RNA structures such as oligonucleotide-based hydrogels or nanoparticles.
Today, bDNA, developed in 1989 by Horn and Urdea, is widely used in clinical and research laboratories for quantitative detection of specific nucleic acid sequences. Assays utilizing bDNA have a wide dynamic range and allow reliable detection of a few target molecules in various samples.
Unlike in PCR, in a bDNA assay, only the signal is amplified, lowering the risk for detecting contaminations. Also, a minor sequence variation in probe-binding regions of the targets does not compromise assay performance, resulting in a more robust quantitation across genotypes. At the lower end of the assay's dynamic range, reproducibility is usually superior to PCR assays.
The synthesis of branched DNA oligonucleotides is possible using protected phosphoramidites with two protected hydroxyl functional groups as the branching monomers. To synthesize branched oligonucleotides used as signal-amplifying multimers, Horn and Urdea utilized standard phosphoramidite chemistry to synthesize a primary oligonucleotide on controlled pore glass (CPG) supports. The scientists used two primary hydroxyl functional groups for the synthesis of branched oligonucleotides on a solid phase. The use of dimethoxytrityl protection or 0-levulinate protection enables the incorporation of the branching points into the oligonucleotide. The synthesis of protected phosphoramidites started from 4-(1,2,4-triazole)-1-(β,-D-5-O- dimethoxytrityl-3-O-t-butyldimethylsilyl-2-deoxyribofuranosyl)-5-methyl-2(1H)- pyrimidinone followed by triazole displacement with 6-aminohexanol.
The researchers used two strategies for the synthesis of bDNAs.
1. The first synthesis strategy utilizing dimethoxytrityl protection for both primary hydroxyl functions resulted in "fork" structures.
2. The second strategy is employing levulinate protection resulted in "comb" structures.
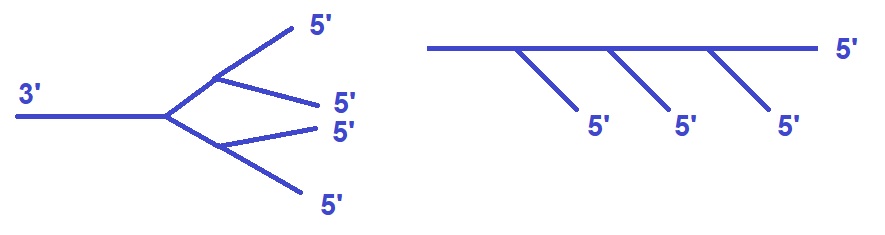
Figure 1: Synthesis strategies for branched DNA (bDNA). (left) Fork structures. (right) Comb structures.
The simultaneous deprotection of the two hydroxyl functions in the branching nucleotides allowed the simultaneous condensation of two additional branching monomers or of two identical nucleoside phosphoramidites to the first oligonucleotide sequence. Deprotection of the functional groups produced two reactive sites that allowed further synthesis. The incorporation of multiple branching monomers permitted the addition of several secondary sequences directly to the primary sequence. After successfully synthesizing the primary sequence, the researchers condensed the oligomer with nucleotide phosphoramidite derivatives that possessed two protected hydroxyl functions.
|
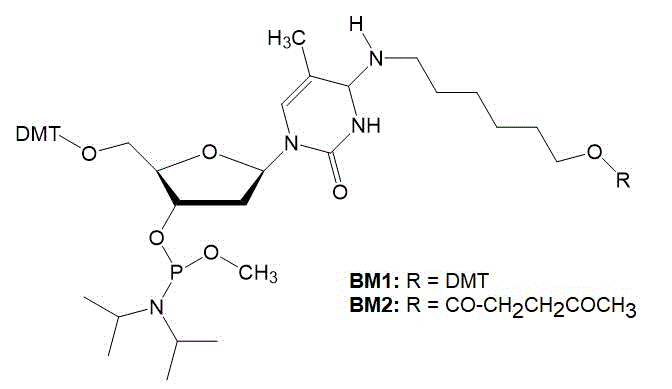
|
Figure 2: Structures of the two phosphoramidites used for the branching monomers (BM1 and BM2).
Horn and Urdea prepared these phosphoramidites to incorporate the branching sites into oligonucleotides.The acid-labile 4,4'-dimethoxytrityl protecting group (DMT) and the levuliniyl protection groups were used to protect the branching hydroxyl group.
The incorporation of nucleoside phosphoramidite derivatives containing two protected primary hydroxyl functional groups into oligonucleotides allowed the formation of fork- or comb-like structures now known as branched oligonucleotides or branched DNA.
|
Fork structure synthesis
The synthesis of the fork structure required synthesis of the primary sequence followed by condensation of one branching nucleotide, two thymidines used as molecular spacers, and two more branching nucleotides, followed by the synthesis of the secondary sequence.
Comb structure synthesis
Using O-levulinate to protect the N4-(6-hydroxylhexyl) functional group allows the synthesis of comb structures. Selective removal of the levulinyl group created reactive groups on the branching nucleotides available for condensing the second sequence or additional branching nucleotides. Two thymidines function as spacers.
Deprotection of the functional groups produced two reactive sites needed for further synthesis. The incorporation of multiple branching monomers allowed the addition of several secondary sequences directly to the primary sequence. After successful synthesis, the researchers condensed the oligomer with nucleotide phosphoramidite derivatives that possessed two protected hydroxyl functions.
The first-generation signal-amplifying assay used large, branched single-stranded polydeoxyribonucleotides (bDNA) combined with a labeled short hybridization probe to amplify targeted nucleic acid sequences.
In subsequent years, Collins et al. developed quantitative hybridization assays based on branched DNA signal amplification further.
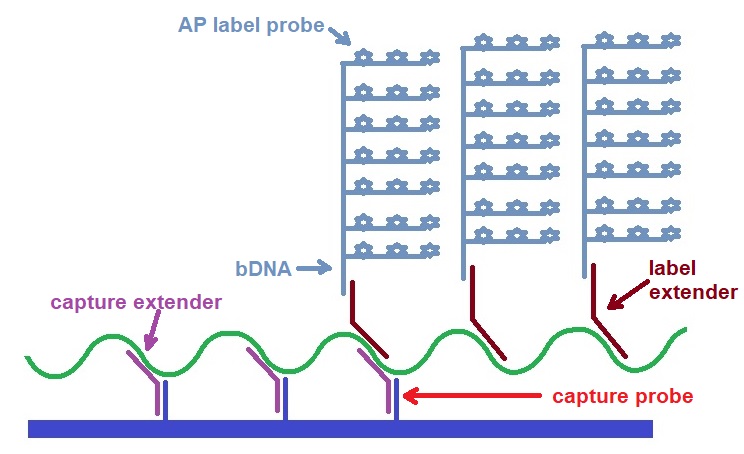
Figure 3: Illustration of the basic first generation bDNA assays showing assay component (Adapted from Collins et al. 1997).
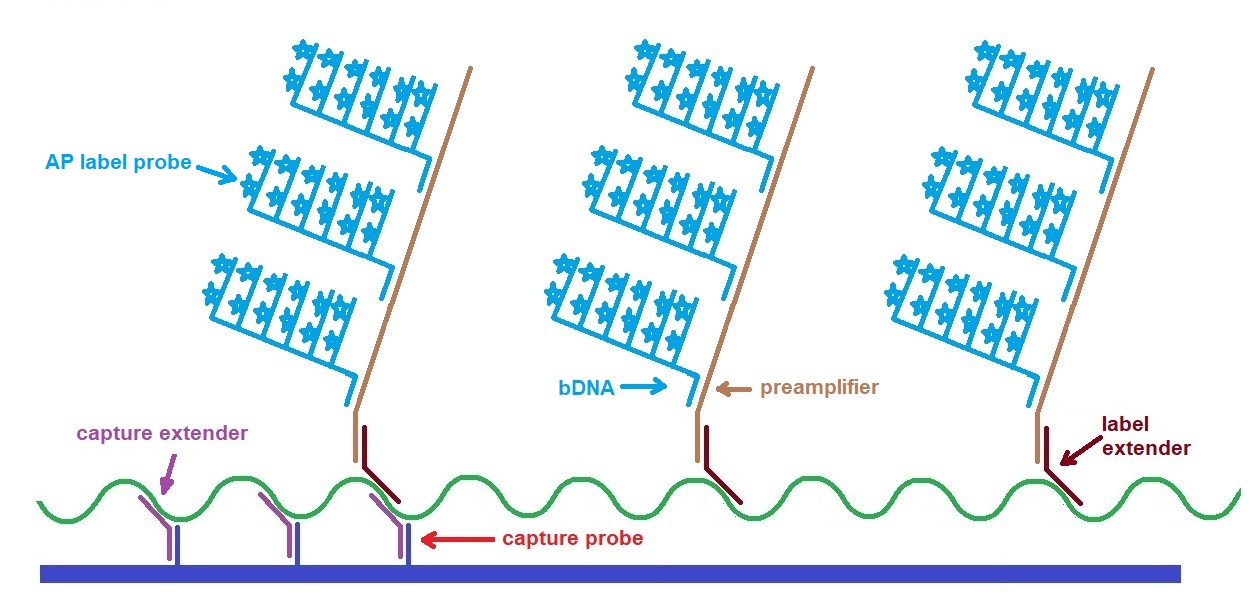
Figure 4: Illustration of the second and third generation bDNA assays showing assay component. This assay approach utilizes preamplifier oligonucleotides (Adapted from Collins et al. 1997).
Oligonucleotides called capture extenders (CEs) are used to capture the target to a solid support. Labeling the target is achieved by binding many target-specific oligonucleotides called label extenders (LEs). In the first generation assay, the LE probes bind a branched DNA amplifier (bDNA) which binds many alkaline phosphatase probes. In the second and third-generation assays, the LE probes bind preamplifiers, which bind many amplifiers, resulting in solid signal amplification and lower detection limits. The linearly amplified signal is directly related to the number of targets present in the original sample.
As descripted by Collins et al., the designed sequences for the preamplifier, amplifier, and alkaline phosphatase probe contain ~30% isobases [5-methyl-2’-deoxyisocytidine (isoC) and 5-methyl-2’-deoxyisoguanosine (isoG)]. Also, the oligonucleotide sequences did not contain any secondary structures longer than a trimer and showed no interactions with any sequence longer than a trimer except with their complements. The targeted melting temperature (Tm) for the oligonucleotides used is above 80 ºC at 1 nM in 3x sodium citrate buffer (NaCl-Na citrate buffer at pH 7; SSC).
Changing the chemistry of the nucleic acid backbone in oligonucleotides will influence their base pairing. The use of alternative or nonstandard base pairs is thought to allow expanding the genetic DNA alphabet. The isoguanine (isoG):isocytosine (isoC) nonstandard base pair forms three H-bonds similar to the guanine (G):cytosine (C) base pair. Because isoC is less stable under alkaline conditions, the 5-methyl derivative (isoCMe) is often used.
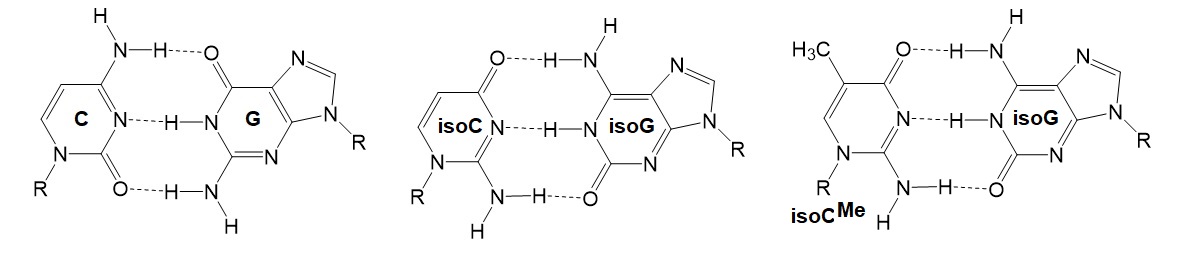
Figure 5: Base-pair motifs for C = G, isoC = isoG, and isoCMe = isoG in antiparallel strand orientation. The corresponding phosophoramidites are useful as building blocks for the synthesis of oligonucleotides containing nonstandard base pairs. Xeno nucleic acids (XNA) also utilize this base pairing to create modified organisms (XMO’s) unable to communicate genetic information with the environment.
Reference
Siti Noor Fathilah Ahmad Ariffin. Mini Review Branched DNA: A Novel Technique for Molecular Diagnostics in Bone Studies. Research Updates in Medical Sciences (RUMeS) 2013, Volume 1; Issue 1 page 27-29.
Collins ML, Zayati C, Detmer JJ, Daly B, Kolberg JA, Cha TA, Irvine BD, Tucker J, Urdea MS. Preparation and characterization of RNA standards for use in quantitative branched DNA hybridization assays. Anal Biochem. 1995 Mar 20;226(1):120–129. [PubMed]
Collins ML, Irvine B, Tyner D, Fine E, Zayati C, Chang C, Horn T, Ahle D, Detmer J, Shen LP, Kolberg J, Bushnell S, Urdea MS, Ho DD. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 1997 Aug 1;25(15):2979-84. [PMC]
Herdewijn P, Marlière P. Toward safe genetically modified organisms through the chemical diversification of nucleic acids. Chem Biodivers. 2009 Jun;6(6):791-808. [PubMed]
Horn T, Urdea M (1989) Forks and combs and DNA: the synthesis of branched oligonucleotides. Nucleic Acids Res 17: 6959–6967. [PMC]
T. Horn, C.A. Chang, and M.S. Urdea; Chemical synthesis and characterization of branched oligodeoxyribonucleotides (bDNA) for use as signal amplifiers in nucleic acid quantification assays. Nucleic Acids Res, 1997, 25, 4842-4849. [PMC]
Kern D, Collins M, Fultz T, Detmer J, Hamren S, Peterkin JJ, Sheridan P, Urdea M, White R, Yeghiazarian T, Todd J. An enhanced-sensitivity branched-DNA assay for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996 Dec;34(12):3196-202. doi: 10.1128/jcm.34.12.3196-3202.1996. [PMC]
5-Me-dC Brancher Phosphoramidite
S, Pezo V, Schepers G, Pinheiro VB, Lescrinier E, Holliger P, Marlière P, Herdewijn P. Isoguanine and 5-methyl-isocytosine bases, in vitro and in vivo. Chemistry. 2015 Mar 23;21(13):5009-22. [PMC]
Urdea, M. Branched DNA Signal Amplification. Nat Biotechnol 12, 926–928 (1994). [nature biotechnology]