The discovery of the epigenetic modification of DNA has been enlightening as it has opened up novel ways of regulating gene expression in normal processes (ex. embryogenesis) as well as in disease progression (ex. carcinogenesis) without changing the genetic code. Likewise, RNA undergoes modification affecting tRNA, rRNA, noncoding RNA (ex. lncRNA), etc. The process of 'RNA editing' was first discovered in 1986 upon identifying nongenomically inserted U in the mitochondrial mRNA of trypanosome, which causes 'sleeping sickness' (Benne et al., 1986). 'Epi-transcriptome' refers to the modifications in mRNA and may affect the expression of transcripts, whose dysfunction is associated with various diseases including cancer.
Presently, more than 150 distinct RNA modifications have been identified (McCowan et al., 2020), which includes 111 for transfer RNAs (tRNAs), 33 for ribosomal RNAs (rRNAs), 11 for noncoding RNAs. The type of RNA modification could be simple (ex. methylations, thiolation, hydroxylation), complex (ex. ring closure, acylation, glycosylation, aminoacylation) or unique (ex. incorporate selenium).
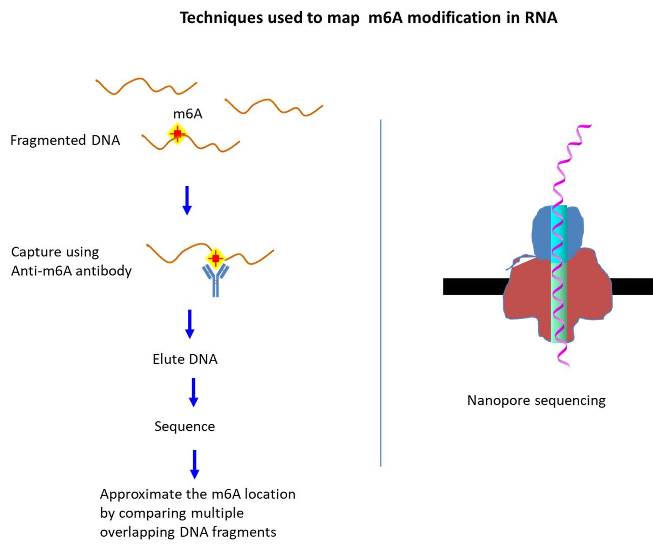
As for the mRNA, 17 modifications have been identified thus far (McCowan et al., 2020). The lesser number of modifications found for mRNA reflects the difficulty of mapping the modified base position precisely and the lack of information regarding the identity of the interacting proteins (Chen et al., 2020). The well known modifications include methylation [ex. N6-methyladenosine (m6A), N1-methyladenosine (m1A), 2-O-dimethyladenosine (m6Am), 5-methylcytosine (m5C)] and isomerization [ex. pseudouridine (Ψ)] (Visser et al., 2020). Among them, m6A represents the most abundant type as it accounts for 0.2-0.5% of total adenosine nucleotides in cellular mRNA (2 or 3 sites per transcript). Initially, however, the prevalence of m6A was not readily accepted, in part, due to the inability to detect, which was greatly improved via the immunoblotting technique (Meyer et al., 2014).
Each type of mRNA modification may vary in terms of distribution as well as functionality. In the case of m6A, which affects ~30% of brain transcripts (also tRNA, rRNA, noncoding RNA), it is commonly added internally near the stop codon or in the 3-UTR (untranslated region). Though it does not affect the specificity of base pairing, the identity of the associating protein (i.e. "reader") dictates the fate of the modified mRNA (ex. YTHDC1 promotes translation, YTHDF2 enhances degradation). In contrast, m6Am (modifies ~0.016% of all adenines in humans) occurs at the first nucleotide (following m7G cap) at the mRNA terminus and may increase stability to enhance expression. The m1A modification, affecting ~20% of transcripts, occurs primarily in the 5'-UTR near the translational initiation site and may increase translation. The m5C addition, which occurs in tRNA, rRNA and mRNA, may interact with the ALYREF protein to promote nuclear export. Pseudouridine, which occurs in noncoding RNA as well as mRNA, may function to weaken the RNA-to-protein interaction (ex. PUF protein) or stabilize the structure of RNA (Vissers et al., 2020).
Mechanistically, multiple means of modifying RNA have been identified. As for the "writer", the modification may be achieved enzymatically, i.e. methyl transferase complex (comprised of METTL3, METTL14, HAKAI, KIAA1429, RBM15/B) for m6A modification, DNMT2 or NSUN2 for m5C modification, PUS1 or PUS7 for isomerization of uridine to pseudouridine (Vissers et al., 2020). The A to I (inosine) deamination is mediated by ADAR (double-stranded RNA-specific adenosine deaminase) (Nishikura, 2019) whereas C to U deamination is mediated by the PPR (pentatricopeptide repeat) protein (Takenaka et al., 2014).
For 2'-O-methylation (rRNA) or the pseudouridine conversion, the modification may be mediated by a distinct complex [consisting of snoRNA (small nucleolar RNA targeting a specific mRNA sequence) and 4 proteins (ex. fibrillarin/Nop1p, NOP56, NOP58, SNU13 for the 'C/D box' snoRNA)]. For the capped mRNA, 2'-O-methylation of first and second nucleotide is catalyzed by CMTR1 (hMTR1) (Belanger et al, 2010) and CMTR2 (hMTR2), respectively (Werner et al, 2011). Similar (but catalytically distinct) methyl transferases are encoded by various viruses (Smietanski et al., 2014).
RNA editing also includes deletion or insertion, and, in the case of negative stranded RNA genome containing paramyxovirus (ex. measles, mumps virus), stuttering of RNA polymerase at a discrete site could lead to G insertion, causing a frameshifting of the open reading frame (Hausemann et al., 1999). In the case of mitochondrial mRNA, the U insertion requires aligning of mRNA to the template 'guide RNA', whose associating proteins then endonucleolytically excise the mRNA to insert U before re-joining (Simpson et al., 1995; Blum et al., 1999).
The role of the mRNA cap structure in avoiding host immune response has been well recognized. In the case of COVID-19 coronavirus, its virion contains 5'-capped genomic RNA with 3'-poly-A tail, which is transcribed into negative strand to serve as a template for generating additional copies of its genome as well as multiple shorter sub-genomic RNAs for protein production. The latter is modified by (1) RNA/NTP triphosphatase (Nsp13; to hydrolyze first phosphate), (2) guanylyltransferase enzyme (Nsp12) to transfer G to form Gppp-RNA, (3) Nsp10/14 to methylate the G at N7 using S-adenosylmethionine, (4) Nsp10/16 for 2'-O-methylation of the first ensuing nucleotide, which is critical to deter recognition by RIG-1 or MDA5 to incite interferon response, whose 3D structure was determined using fixed-target serial synchrotron crystallography (Wilamowski et al., 2021; Basu et al., 2021).
The key to preventing epidemic is the ability to diagnose the infected early to preempt further propagation. For this, Bio-Synthesis, Inc. provides primers and probes (as well as synthetic RNA control) for COVID-19 diagnosis via RT-PCR assay. It specializes in oligonucleotide modification and provides an extensive array of chemically modified nucleoside analogues (over ~200) including bridged nucleic acid (BNA) in addition to mRNA synthesis. A number of options are available to label oligonucleotides (DNA or RNA) with fluorophores either terminally or internally as well as to conjugate to peptides or antibodies. It recently acquired a license from BNA Inc. of Osaka, Japan, for the manufacturing and distribution of BNANC, a third generation of BNA oligonucleotides. To meet the demands of therapeutic application, its oligonucleotide products are approaching GMP grade. Bio-Synthesis, Inc. has recently entered into collaborative agreement with Bind Therapeutics, Inc. to synthesize miR-21 blocker using BNA for triple negative breast cancer. The BNA technology provides superior, unequalled advantages in base stacking, binding affinity, aqueous solubility and nuclease resistance. It also improves the formation of duplexes and triplexes by reducing the repulsion between the negatively charged phosphates of the oligonucleotide backbone. Its single-mismatch discriminating power is especially useful for diagnosis (ex. FISH using DNA probe). For clinical application, BNA oligonucleotide exhibits lesser toxicity than other modified nucleotides.
https://www.biosyn.com/oligo-flourescent-labeling.aspx
https://www.biosyn.com/tew/Speed-up-Identification-of-COVID19.aspx
https://www.biosyn.com/covid-19.aspx
https://www.biosyn.com/mrna.aspx
https://www.biosyn.com/tew/Messenger-RNA-turnover-and-their-half-live.aspx#!
https://www.biosyn.com/tew/Pseudouridine,-an-abundant-post-transcriptional-RNA-modification.aspx
References
Basu S, Mak T, et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of Nsp14 RNA cap methyltransferase. Biochem J. 478:2481-2497 (2021). PMID: 34198328
Benne, R., J. Van den Burg, et al. Transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46:819–826 (1986). PMID: 3019552
Bélanger F, Stepinski J, et al. Characterization of hMTr1, a human Cap1 2'-O-ribose methyltransferase. J Biol Chem. 285:33037-33044 (2010). PMID: 20713356
Blum B, Bakalara N, et al. A model for RNA editing in kinetoplastid mitochondria: "guide" RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 60:189-98 (1990). PMID: 1688737
Chen LQ, Zhao WS, et al. Mapping and editing of nucleic acid modifications. Comput Struct Biotechnol J. 18:661-667 (2020). PMID: 32257049
Hausmann S, Law FM, et al. Regulation of parathyroid hormone/parathyroid hormone-related protein receptor expression by osteoblast-deposited extracellular matrix in a human osteoblast-like cell line. J Cell Physiol. 165:164-71 (1995). PMID: 7559797
McCown PJ, Ruszkowska A, et al. Naturally occurring modified ribonucleosides. Wiley Interdiscip Rev RNA. 11:e1595 (2020). PMID: 32301288
Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 15:313-26 (2014). PMID: 24713629
Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 79:321-49 (2010). PMID: 20192758
Simpson L, Thiemann OH. Sense from nonsense: RNA editing in mitochondria of kinetoplastid protozoa and slime molds. Cell. 81: 837–40 (1995). PMID: 7781060
Smietanski M, Werner M, et al. Structural analysis of human 2'-O-ribose methyltransferases involved in mRNA cap structure formation. Nat Commun. 5:3004 (2014). PMID: 24402442
Takenaka M, Verbitskiy D, et al. RNA editing in plant mitochondria-connecting RNA target sequences and acting proteins. Mitochondrion. Pt B:191-7 (2014). PMID: 24732437
Vissers C, Sinha A, et al. The epitranscriptome in stem cell biology and neural development. Neurobiol Dis. 146:105139 (2020). PMID: 33065280
Werner M, Purta E, et al. 2'-O-ribose methylation of cap2 in human: function and evolution in a horizontally mobile family. Nucleic Acids Res. 39:4756-68 (2011). PMID: 21310715
Wilamowski M, Sherrell DA, et al. 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography. Proc Natl Acad Sci U S A. 118:e2100170118 (2021). PMID: 33972410