Off-target effects in RNA interference screens occur where small interfering RNAs directly affect the expression of genes other than the targeted gene. Microarray experiments indicated that small interfering RNAs (siRNA) could affect the mRNA levels of many genes.
The recent approval of multiple siRNA therapeutics highlights the potential of siRNA drugs. The success of siRNA drugs was possible due to the availability of chemically modified nucleic acids and their impact on the stability, delivery, potency, and off-target effects of siRNAs.
Therapeutic small interfering RNAs (siRNAs) are double-stranded RNAs that specifically silence genes through the activation of the RNA interference pathway (RNAi). Double-stranded RNAs allow the design of potent siRNAs for specific gene silencing. Since siRNAs are delivered nakedly, they must be modified to resist degradation. Each siRNA strand can downregulate several genes. Also, a saturation of the RNAi machinery leads to the upregulation of miRNA-repressed genes. Often clinical trials face failure because of off-target effects such as off-target gene dysregulation.
Eliminating sense strand uptake reduces off-target gene silencing and limits the disruption to endogenous regulatory mechanisms. Therefore, optimal strand selection impacts the success of future siRNA therapeutics. Extensive modification and encapsulation in protective carrier molecules can prevent the rapid degradation by blood serum nucleases of siRNAs. The formulation in lipid nanoparticles (LNPs) facilitates the delivery of siRNAs to tissues such as the liver. The chemical conjugation of double-stranded siRNAs to biological molecules such as GalNac enables their cellular delivery.
Cuccato et al. estimated that 103 to 104 RISC molecules are present in the average mammalian cell, corresponding to an intracellular concentration of approximately 3 to 5 nM.
Each siRNA strand can have a range of distinct gene-silencing effects. Limiting sense strand selection allows the management of off-target effects. Minimizing off-target effects is possible using chemical modification and sequence design strategies which reduce seed-region binding and possible matches in the 3’-untranslated region (3′UTR).
Conjugates can inhibit their corresponding strands resulting in the development of conjugates added to the sense strand.
siRNAs modified with hydrophobic conjugates and adding 5’-E-vinylphosphonate (5′-E-VP) improves tissue retention, silencing activity, and duration of the silencing effect.
Nomenclature
In the siRNA nomenclature as proposed by Varley & Desaulniers in 2020 [AS/SS:##] indicates the duplex molecules where AS refers to the antisense strand, SS refers to the sense strand, and ## indicates the nucleotide number from the 5′ end of the corresponding strand. For example, SS:14 refers to the 14th nucleotide from the 5′ end of the sense strand.
Chemical modifications of siRNA impacting strand selection.
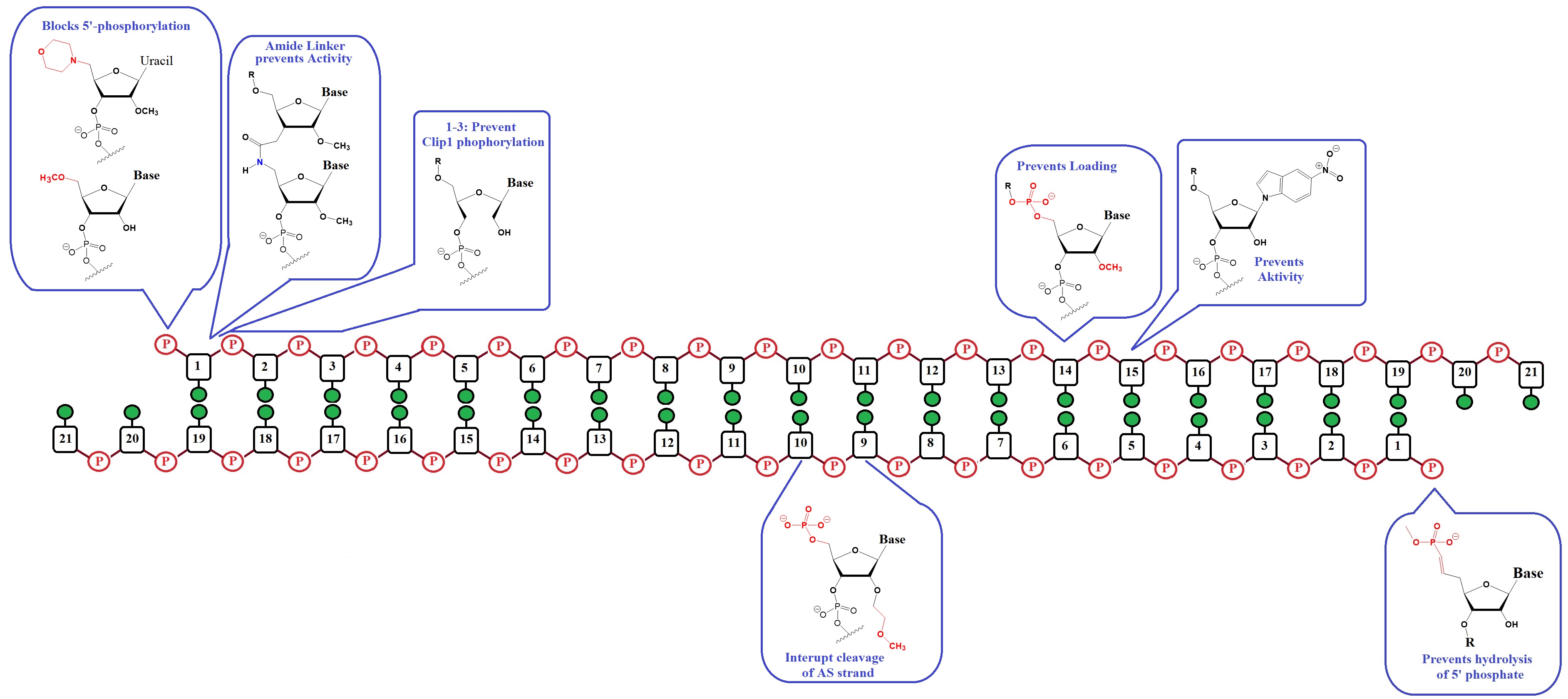
Figure 1: Strategies to mitigate sense strand uptake into RISC (Adapted from Varley & Desaulniers).
On-target and off-target effects:
[1] The 5’-phosphate is the most critical region for the uptake by AGO2.
[2] A phosphate group at the 5’-end of a strand is essential for AGO2 uptake. Clp1 kinase and phosphatase(s), which balance 5’-phosphorylated and free hydroxyl ends in cells, recognize siRNAs.
[3] The efficiency of these enzymes is influenced by modifications of the 5'-ends of these enzymes resulting in a shift in phosphorylation which affects the likelihood of uptake of the associated strand.
Effectively stabilized 5’-phosphate mimics must facilitate RNAi activity by overcoming three hurdles:
[1] They must sterically fit within the deep binding pocket of AGO2;
[2] Permit interactions with AGO2 side chains without significant distortion to the attached nucleoside; and
[3] Prevent hydrolysis by phosphatases within the cell.
These strict requirements resulted in relatively few effective 5’-phosphate mimics.
Stabilized 5’-phophate mimics for the antisense strand can carry modifications within the first three nucleotides of the 5’-end. These modifications can reduce Clp1 kinase activity, preventing intracellular phosphorylation of the strand.
Successful chemical strategies for improving strand selection for siRNAs are:
[1] Control of the 5’-phosphorylation status.
[2] Alter internal structure to prevent cleavage of the antisense loaded in the passenger strand.
[3] Alter siRNA structure that prevents sense strand capture.
All improvements will support the development of effective, safe, and potent therapeutics.
Kobayashi et al. recently reported rules for siRNA sequences allowing them to be functional in mammalian cells:
[1] Place A/U at the 5′ end of the guide strand, G/C at the 5′ end of the passenger strand. Place more than 4 A/Us in the 5′-terminus 7-nucleotide of the guide strand, and no GC stretch longer than 8 nucleotides.
[2] siRNA sequences with weak off-target activity: siRNAs with low Tm value in the seed region (nucleotides 2–8) exhibit weak off-target effect.
[3] Nucleotides in the seed region can be functionally divided into two domains in response to 2′-OMe modifications:
[a] 2′-OMe modifications of nucleotides 2–5 inhibit the off-target effect of siRNAs
[b] 2′-OMe modifications of nucleotides 6–8 promote both on-target and off-target effects.
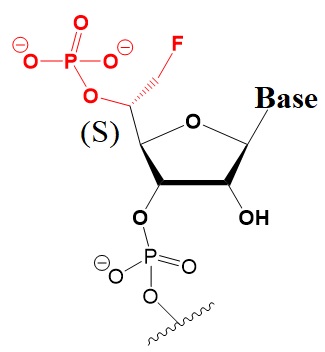
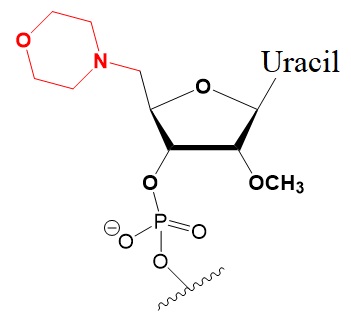
Chemical modifications of the 5’-terminus of siRNA influencing strand selection and antisense activity.
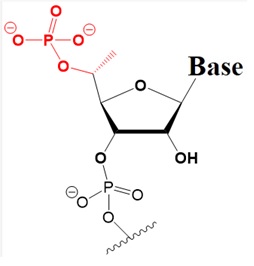
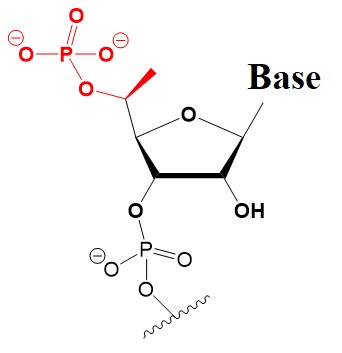
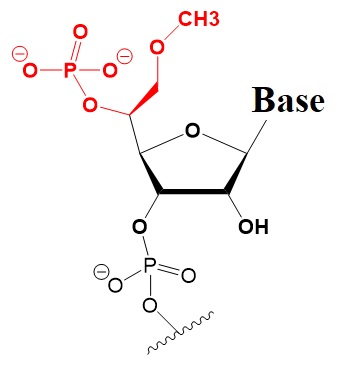
Many researchers have investigated 5’-terminal modifications in siRNA and ss-siRNA. 5’-Phosphate Modifications include methylene- and vinyl-phosphonate, 5′-Hydrogen substitutions, and 5′-alternative structures. The structures of these modifications are shown below (Varley & Desaulniers, 2021).
|
.jpg)
|
|
|
Natural 5’-Phosphate
|
|
|
.jpg)
|
.jpg)
|
|
|
|
5-Methylene Phosphonate NA.
|
5-(E)-Vinyl-Phosphonate NA.
5′-(E)-VP phosphate mimic is the preferred choice for phosphate stabilization.
Addition to a GalNac3 conjugated siRNA improves the IC50 3 to 20 fold.
|
5-(R)-Me-Phosphonate NA.
Improves the IC50 ~ 2 to 3-fold.
|
5-(S)-Me-Phosphonate NA.
No impact on IC50 of siRNA.
|
|
|
|
|
.jpg)
|
|
5-R-MeOCH3-Phosphonate NA.
Improves activity 2 to 20-fold.
|
5’-(S)-FMe-P NA.
Improved IC50 ~ 2-fold.
|
5’-Deoxy-5’-Morpholino-2’-O-Methyl Uridine.
Reduced activity ~3-fold.
May prevent uptake of sense strand.
|
5’-O-Methyl NA.
Disrupts activity.
|
|
|
|
|
|
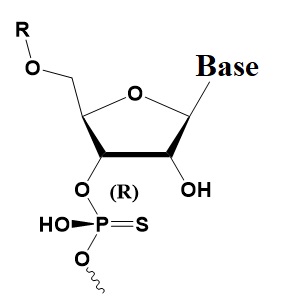
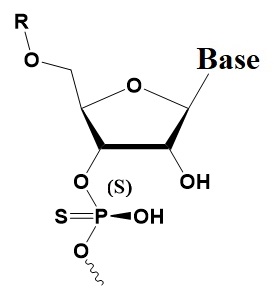
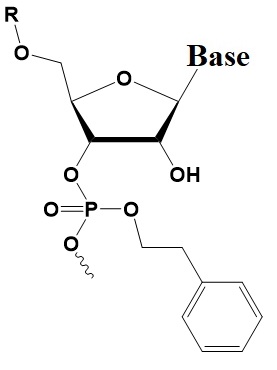
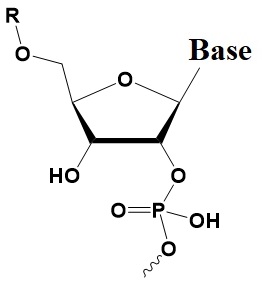
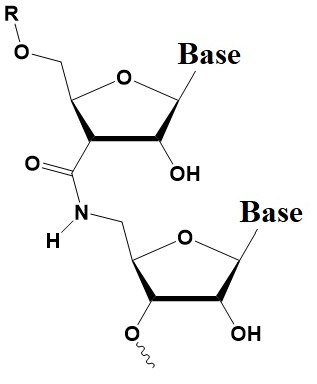
Chemical internal modifications of siRNA influencing strand selection and antisense activity.
|
|
|
|
|
|
|
Rp Phosphorothioate
|
Sp Phosphorothioate
|
Phenylethyl Phosphate
|
2’-5’-Phosphate linkage
|
Amide linkage
|
Reference
Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, Marshall WS, Khvorova A: 3' UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006, 3 (3): 199-204. [Nature Methods]
Chernikov IV, Gladkikh DV, Meschaninova MI, Ven'yaminova AG, Zenkova MA, Vlassov VV, Chernolovskaya EL. Cholesterol-Containing Nuclease-Resistant siRNA Accumulates in Tumors in a Carrier-free Mode and Silences MDR1 Gene. Mol Ther Nucleic Acids. 2017 Mar 17;6:209-220. [PMC]
Cuccato G, Polynikis A, Siciliano V, Graziano M, di Bernardo M, di Bernardo D. Modeling RNA interference in mammalian cells. BMC Syst Biol. 2011 Jan 27;5:19. [PMC]
Dong Y, Siegwart DJ, Anderson DG. Strategies, design, and chemistry in siRNA delivery systems. Adv Drug Deliv Rev. 2019 Apr;144:133-147 [PMC]
Fedorov Y, Anderson EM, Birmingham A, Reynolds A, Karpilow J, Robinson K, Leake D, Marshall WS, Khvorova A. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12(7):1188–1196. [PMC]
Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS: Widespread siRNA "off-target" transcript silencing mediated by seed region sequence complementarity. RNA. 2006, 12 (7): 1179-1187. [PMC]
Varley AJ, Desaulniers JP. Chemical strategies for strand selection in short-interfering RNAs. RSC Adv. 2021 Jan 11;11(4):2415-2426. [PMC]
---...---
" Bio-Synthesis provides a full spectrum of high quality custom oligonucleotide modification services including back-bone modifications, conjugation to fatty acids, biotinylation by direct solid-phase chemical synthesis or enzyme-assisted approaches to obtain artificially modified oligonucleotides, such as BNA antisense oligonucleotides, mRNAs or siRNAs, containing a natural or modified backbone, as well as base, sugar and internucleotide linkages.
Bio-Synthesis also provides biotinylated and capped mRNA as well as long circular oligonucleotides".
---...---