Molecular probes containing bridged nucleic acids (BNAs) at selected positions in combination with real-time PCR allow the detection of pathogen mutants that can cause various disorders in humans.
Nontuberculous mycobacteria are such pathogens. Nontuberculous mycobacteria (NTM) are mycobacteria other than M. tuberculosis and M. leprae. NTM are known as atypical mycobacteria, mycobacteria other than tuberculosis (MOTT), or environmental mycobacteria.
Hirama et al. designed a BNA probe detection system based on real-time PCR (called BNA-PCR) to identify point mutations at position 2058 or 2059 in domain V of the 23S rRNA gene responsible for clarithromycin resistance.
BNA are artificial nucleic acids with a high binding affinity to specific sequences allowing the design of probes with improved hybridization capability and enhanced biochemical stability. Specific designed BNA probes enable discrimination of single mismatches within a target sequence of amplified products.
The BNA-PCR assay allows the identification of Mycobacterium avium complex (MAC) isolates from clinical samples.
Clarithromycin resistance gene BNA-PCR primer and probe set
Forward primer: 5′-GTAACGACTTCCCAACTGTCTC-3′
Reverse Primer: 5′-ACCTATCCTACACAAACCGTACC-3′
BNA Probe: Alex532-CGCGGCAGGACGAAAAGAC-BHQ1 ; AA = Placing of BNAs
The BNA FRET probe contains two BNA bases for detecting MAC isolates' clarithromycin resistance gene (BNAs are colored). If a point mutation is present, the resulting sequence mismatch leads to a decrease in the melting temperature (Tm) value, preventing the BNA probe from annealing to the PCR product, resulting in a reduced emitted fluorescence (Hirama et al.).
Recently Bio-Synthesis also developed an “Ultra-Sensitive BRAF Codon 600 (V600E) Mutation Analysis Kit” based on BNAs for rapid and convenient real-time PCR detection of the BRAF-V600E mutation with high sensitivity. The kit enables efficient discrimination of the mutant from the wild-type gene. It allows laboratories to detect levels down to 0.1% or lower of the BRAF V600E mutant in a wild-type background.
According to the CDC, the opportunistic pathogen NTM infects people more quickly with underlying lung disease or depressed immune systems. However, these pathogens are typically not transmitted person-to-person. NTM is found in soil, dust, and water, including lakes, rivers, and streams, in drinking water and water used to shower. NTM forms difficult-to-eliminate biofilms. Biofilms are a collection of microorganisms sticking to each other and do adhere to surfaces in moist environments, for example, the insides of plumbing in buildings. NTMs can infect many body sites, most commonly the lungs, skin, and soft tissue. However, NTMs are also often found in devices associated with infections, lymph nodes, blood, or other usually sterile locations in the body. Immunocompromised patients, such as those with HIV or AIDS, often have NTM infections.
MAC consist of two Mycobacterium species, M. avium, and M. intracellulare. This infection also causes respiratory illness in birds, pigs, and humans, especially in immunocompromised people. MAC cannot be distinguished in a microbiology laboratory and requires genetic testing.
Mac has been known now for about a hundred years. MAC causes progressive lung disease. Recommended treatment regimens include a macrolide and a rifamycin. However, drug intolerance and relapse after completed treatment often limit a successful therapy. Clarithromycin is the most efficacious drug among the various treatment regimens for lung NTM.
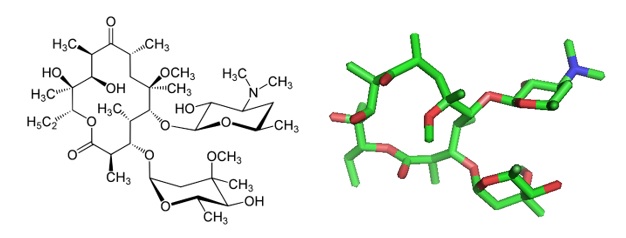
Figure 1: Clarithromycin, structures. Clarithromycin is a ribosome-targeting macrolide antibiotic that interacts with a bacterial 23S rRNA hairpin-like structure in domain II and the peptidyl transferase loop in domain V.
Reference
23S rRNA
Alexa 532 labeling
BHQ1 modification
BNA Clamp PCR
Bridged Nucleic Acids (BNAs)
CDC nontuberculous-mycobacteria
Clarithromycin
FRET molecular beacon
Dye and Quencher Pairs for FRET
Sung-Kun Kim, Klaus D. Linse, Parker Retes, Patrick Castro, Miguel Castro; Bridged Nucleic Acids (BNAs) as Molecular Tools.J Biochem Mol Biol Res 2015 September 1(3): 67-71. [PDF]
Hirama T, Shiono A, Egashira H, Kishi E, Hagiwara K, Nakamura H, Kanazawa M, Nagata M. PCR-Based Rapid Identification System Using Bridged Nucleic Acids for Detection of Clarithromycin-Resistant Mycobacterium avium-M. intracellulare Complex Isolates. J Clin Microbiol. 2016 Mar;54(3):699-704. [PMC]
Imanishi T, Obika S. 2002. BNAs: novel nucleic acid analogs with a bridged sugar moiety. Chem Commun (Camb.) 16:1653–1659. [ Article ]
Rahman SA, Seki S. 2008. Design, synthesis, and properties of 2′,4′-BNANC: a bridged nucleic acid analogue. J Am Chem Soc 130:4886–4896. [PDF]
“Ultra-Sensitive BRAF Codon 600 (V600E) Mutation Analysis Kit” based on BNAs.
---...---