MicroRNAs or miRNAs are a class of endogenous small RNAs approximately 22 nucleotides in size found in plants and animals including humans. More and more research data indicate that in humans microRNAs or miRNAs are useful indicator molecules for different cancer types which may be useful as cancer biomarkers.
Increasingly, it has become apparent that microRNAs take part in the development of cancer. This observation has made miRNAs potential biomarkers for cancer diagnosis and prognosis. Therefore it can be reasoned that microRNAs or miRNAs are good candidates as cancer biomarker. Because many studies now suggest that the pattern of microRNA expression in tissues reflects the disease status in this tissue, miRNA expression levels may serve as potential biomarkers with multiple applications in clinical diagnostics. miRNAs can be successfully isolated from biological fluids allowing for the development of biofluid biopsies or diagnostics. This type of biomarker diagnostics promises to allow for the development of minimal invasive assays, to save cost and simplify complex invasive procedures.
miRNA's processing occurs from approximately 70 nucleotides in size hairpin precursor RNAs by the protein Dicer. miRNA have been shown to regulate their target messengerRNA (mRNA) by destabilizing mRNA molecules and translational repression.
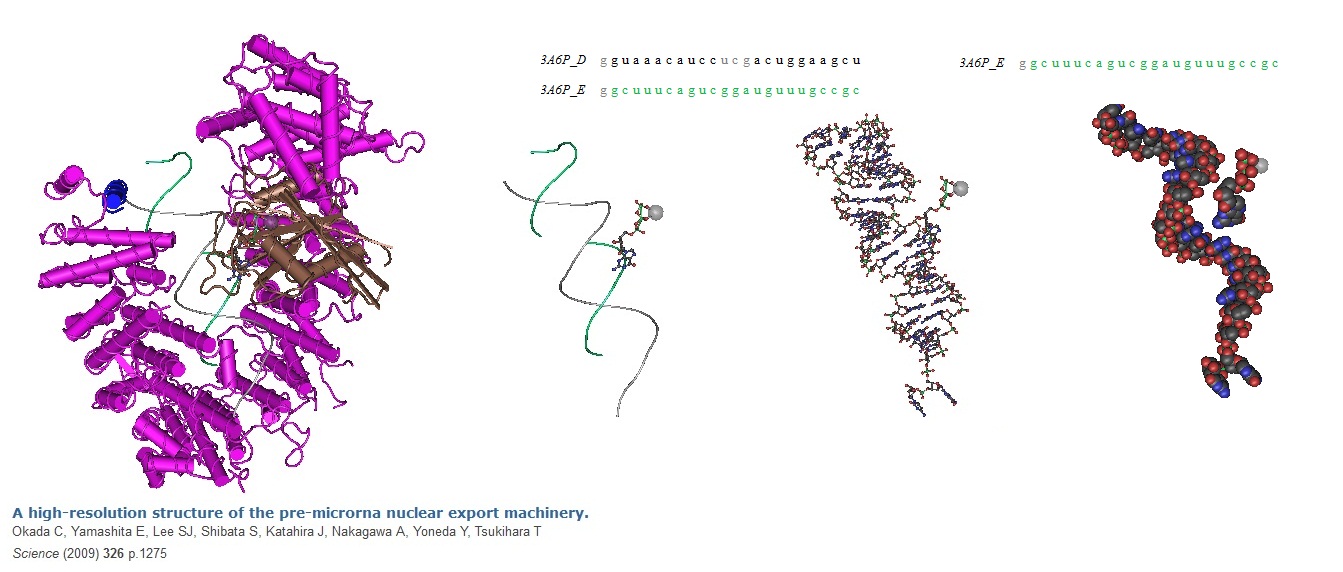
Figure 1: Pre-miRNA nuclear export machinery.
Okada et al in 2009. solved the structure of the "pre-miRNA nuclear export machinery" formed by pre-miRNA complexed with Exp-5 and a guanine triphosphate (GTP)-bound form of the small nuclear guanine triphosphatase (GTPase) Ran (RanGTP) at 2.9 angstrom. The data showed that RNA recognition by Exp-5:RanGTP does not depend on RNA sequence. This implys that Exp-5:RanGTP can recognize a variety of pre-miRNAs.
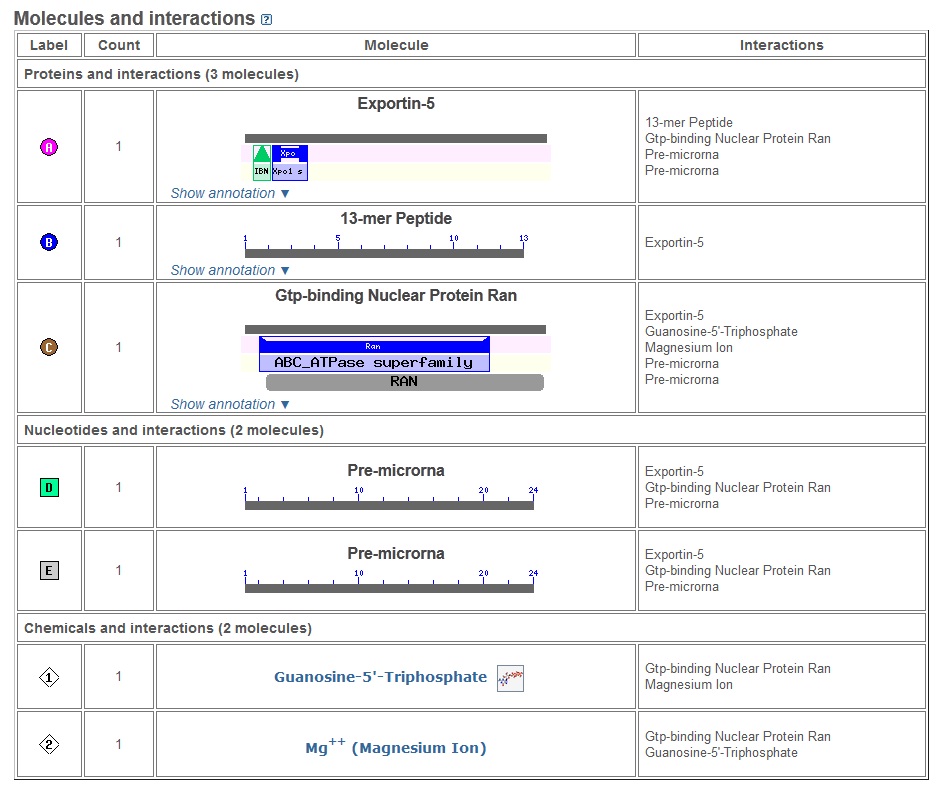
Figure 2: The molecules present in the structure and their interactions are shown here.
[Source: http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=78532]
During a study of the nematode Caenorhabditis elegans (C. elegans) development involving the gene lin-14 Victor Ambros, Rosalind Lee and Rhonda Feinbaum first discovered miRNAs in 1993. However, at the time the researcher speculated that these molecules could be a nematode idiosyncrasy. In 2000, it was shown that let-7 represses lin-41, lin-14, lin28, lin42 and daf12 mRNA during transition in developmental stages in C. elegans. At this time miRNAs were recognized as small regulatory RNAs. Furthermore, it became clear that miRNAs are conserved in many species. In addition, it was noted that short non-coding RNAs, first identified in 1993, were part of a wider phenomenon. For example, Lagos-Quintana et al. in 2001 referred to 22- and 21-nucleotide (nt) RNAs as small temporal RNAs (stRNAs). These RNAs functioned as key regulators in developmental timing. The Tuschl lab in 2001 showed that many 21- and 22-nt expressed RNAs exist in invertebrates and vertebrates. Furthermore, some of these RNAs, similar to let-7 stRNA, are highly conserved. This discovery led to the conclusion that sequence-specific, posttranscriptional regulatory mechanisms as mediated by small RNAs are more general than was previously appreciated. Over 4000 miRNAs have been found so far in all studied eukaryotes. More than 700 miRNAs have already been identified in humans. In addition, more than and over 800 are predicted to exist. V. Ambros in 2001 reported that these microRNAs are diverse in sequence and expression patterns. The observation that these molecules are evolutionarily widespread suggests that they may participate in a wide range of genetic regulatory pathways. Figure 3 shows the dramatic increase in publications involving miRNAs and miRNA research.
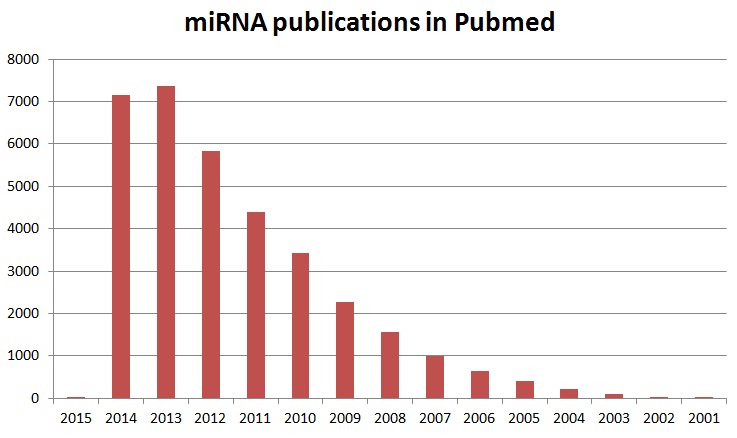
Figure 3: Increase in miRNA publications in Pubmed.
Animal miRNAs derived from longer primary transcripts carry hairpin structures. The processing of these precursor hairpin RNA structures proceeds in a stepwise fashion catalyzed by the RNase III enzymes Drosha and Dicer. Drosha cleaves these RNA molecules near the hairpin base to release the pre-miRNA hairpin. This reaction occurs in the nucleus. Next, the pre-miRNA hairpin is exported into the cytoplasm and Dicer cleaves on the loop side of the hairpin. The result is a miRNA:miRNA* duplex. In the next step, one strand of this complex is preferentially incorporated into a silencing complex.
Recently an alternative nuclear pathway for miRNA biogenesis was identified in invertebrates. Researchers found that short introns with hairpin potential, termed mirtrons, can be spliced and debranched into pre-miRNA hairpin mimics that appear to bypass Drosha cleavage. Debranched mirtrons access the canonical miRNA pathway during nuclear export and are then cleaved by Dicer and incorporated into silencing complexes. As pointed out by Brezikow et al. in 2007, mirtrons are alternative precursor molecules for microRNA biogenesis present in invertebrates. Splicing allows these short hairpin introns to bypass Drosha cleavage. Drosha cleavage is essential for the generation of canonical animal microRNAs. With the help of computational and experimental strategies Brezikow et al. establish that mammals have mirtrons as well. Therefore, mirtrons are miRNAs located in the introns of mRNA encoding genes. Brezikow et al. identified three (3) well conserved mirtrons expressed in diverse mammals. In addition, 16 primate-specific mirtrons, and 46 mirtron candidates, as supported by limited cloning, are suspected to be present in primates as well.