The incorporation of N1-methyl pseudouridine by T7 bacteriophage derived RNA polymerase was instrumental for COVID-19 mRNA vaccine production
For the ongoing pandemic, the mRNA vaccines targeting the spike protein of the COVID-19 coronavirus were introduced in a timely manner to curtail its progression. This preventive measure relies on the ability of the injected mRNA to mount both the humoral and cellular immunity to preempt the oncoming infection. The administration of the mRNA vaccines, which began in the early months of 2021 led to a significant decline in the rate of ensuing COVID-19 infections (though it tends to decline during warmer months). Nevertheless, targeting a single molecular entity (spike protein) of a virus comprised of nearly 29 proteins could be challenging--especially, in lieu of the higher mutation rates of RNA viruses. Hence, the ability of the vaccine to counter the emerging COVID-19 variants is continually being assessed. Supporting the therapeutic value of the mRNA vaccines is the finding that the rate of infection /hospitalization was significantly reduced amongst the vaccinated.
For a considerable period, the ability to synthesize nucleic acids has played a pivotal role in biological and medical researches. It began with the ability to synthesize short oligonucleotides as they could be used in multiple biological applications such as mutagenesis, molecular cloning, DNA sequencing, etc. that involve hybridization. Equally significant is the ability to utilize oligonucleotides for various medical applications including PCR-based assays for diagnosing the disease onset, testing for genetic counseling, monitoring altered gene expression via microarrays, etc. For translational medicine, the potential use of oligonucleotides for therapeutic purposes is increasingly being recognized. This has resulted in numerous ongoing clinical trials and the FDA approval of multiple siRNAs, antisense oligonucleotides, and gapmers. Continuing with the advance is the application of oligonucleotides for gene synthesis as part of the genome construction endeavor at the DNA level.
At the RNA level, the therapeutic potential of the synthetic nucleic acids was extended to mRNAs, which are significantly longer than oligonucleotides. The earlier attempts were to intravenously or intradermally inject melanoma patients with dendritic cells, which have been electroporated with mRNA vaccine encoding cancer immunogens (Wilgenhof et al., 2013). In 2021, the medical utility of the mRNA vaccine technology was tested against the newly emerged COVID-19 coronavirus. To target the spike protein of COVID-19, it required synthesizing mRNAs of >4 kilobases in length. To achieve this length, biopharmaceutical industries resorted to the in vitro transcription technology.
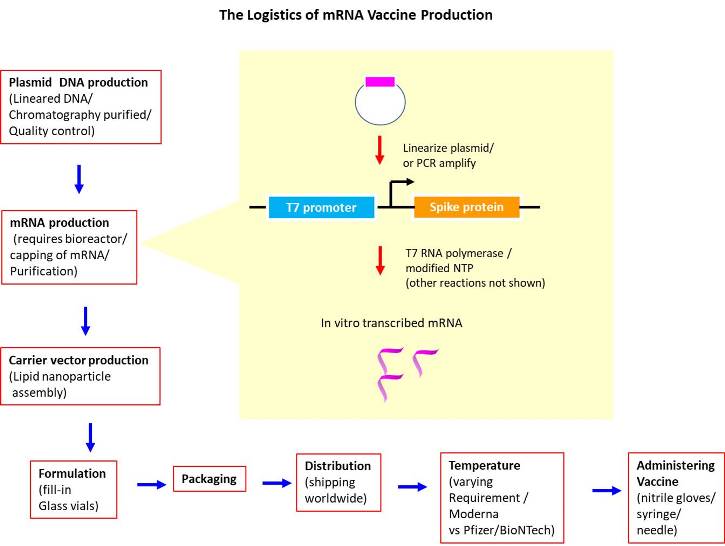
The general design of the template used to produce mRNA vaccines utilized the following format: 5'-cap (to increase translation, stability; to decrease immunogenicity, degradation), 5'-UTR (to promote interaction with ribosome), codon-optimized spike protein coding sequence (to augment translation), 3'-UTR (to enhance stability), and poly-A tail (to increase stability, translation). The mRNA was engineered to incorporate modified nucleic acids to avoid immunogenicity or degradation, ex. N1-methyl pseudouridine (Nance et al. 2021). As such, critical to its development was the the employing of a RNA polymerase that could accommodate nucleotides with non-natural bases.
Generally, there are 3 strategies for the production of RNA with modified nucleotides (Milisavljevič et al., 2018). First approach is to exploit the catalysis of RNA polymerase to incorporate modified NTPs (as in the case of T7 polymerase). The second method is to modify post-transcriptionally using enzymes (ex. using methyl transferase) after the RNA synthesis. Third option is through chemical modification of the RNA using bioothogonal (ex. click) chemistry (George et al.., 2017; Holstein et al., 2016; George et al., 2020).
The ability of RNA polymerase to incorporate modified nucleotides was recognized as early as 1963, when the ability of HeLa cell derived RNA polymerase to incorporate pseudouridine (incorporated adjacent to purine nucleotides preferentially) was observed (Goldberg et al, 1963). Later, the ability of T7 polymerase to incorporate modified NTPs was described, so long as it does not perturb base-pairing (Milligan et al., 1989). Subsequently, it was found that T7 RNA polymerase permits a wide variety of modifications to nucleotides including biotin (affinity labeling), alkyne/azido groups (click chemistry), 5-vinylU (for further chemical modification), 5-iodoU (for cross-coupling modification), amino acid-like side chain (aptamer selection), diazirin (crosslinking), and fluorophore (Milisavljevič et al., 2018). T7 RNA polymerase could also tolerate sugar modifications (ex. 2'-O-carbamoyl uridine) (Masaki et al., 2016; Huang et al., 1997)
Several advances preceded the production of the mRNA vaccine for COVID-19. The bulk synthesis of modified mRNA relied on the cloning/sequencing of T7 bacteriophage gene encoding RNA polymerase by W. Studier and colleagues (Brookhaven National Laboratory, USA) in 1983 (Davaloo et al., 1984). Further attempts to mutagenize T7 RNA polymerase to obtain variants that could incorporate modified nucleotides (ex. 2'-C-branched uridine) also took place (Siegmund et al, 2012; Pavey et al., 2004). Critical was the finding by Kariko and colleagues (University of Pennsylvania, USA) in 2005 that nucleotide modifications such as pseudouridine, thiouridine, and 5-methylcytidine decrease immunostimulation by TLRs (toll-like receptors), as it provided foundation for applying mRNA for anti-COVID-19 vaccine (or stem cell technology, cancer therapy potentially) (Nance et al., 2021; Kariko et al., 2005). The topics of further interest include the mRNA design (ex. the effect of ubiquitous incorporation of N1-methyl pseudouridine on 5'-UTR or 3'-UTR functionality), the impact on diagnosis (Rt-PCR), and recombination.
The key to preventing epidemic is the ability to diagnose the infected early to preempt further propagation. For this, Bio-Synthesis, Inc. provides primers and probes (as well as synthetic RNA control) for COVID-19 diagnosis via RT-PCR assay. It specializes in oligonucleotide modification and provides an extensive array of chemically modified nucleoside analogues (over ~200) including bridged nucleic acid (BNA) in addition to mRNA synthesis. A number of options are available to label oligonucleotides (DNA or RNA) with fluorophores either terminally or internally as well as to conjugate to peptides or antibodies. It recently acquired a license from BNA Inc. of Osaka, Japan, for the manufacturing and distribution of BNANC, a third generation of BNA oligonucleotides. To meet the demands of therapeutic application, its oligonucleotide products are approaching GMP grade. Bio-Synthesis, Inc. has recently entered into collaborative agreement with Bind Therapeutics, Inc. to synthesize miR-21 blocker using BNA for triple negative breast cancer. The BNA technology provides superior, unequalled advantages in base stacking, binding affinity, aqueous solubility and nuclease resistance. It also improves the formation of duplexes and triplexes by reducing the repulsion between the negatively charged phosphates of the oligonucleotide backbone. Its single-mismatch discriminating power is especially useful for diagnosis (ex. FISH using DNA probe). For clinical application, BNA oligonucleotide exhibits lesser toxicity than other modified nucleotides.
https://www.biosyn.com/oligo-flourescent-labeling.aspx
https://www.biosyn.com/tew/Speed-up-Identification-of-COVID19.aspx
https://www.biosyn.com/covid-19.aspx
https://www.biosyn.com/mrna.aspx
https://www.biosyn.com/tew/Messenger-RNA-turnover-and-their-half-live.aspx#!
https://www.biosyn.com/tew/Pseudouridine,-an-abundant-post-transcriptional-RNA-modification.aspx
https://www.biosyn.com/tew/the-potential-use-of-dna-or-mrna-based-vaccines-incorporating-modified-nucleotides-to-suppress-cancer-or-covid-19-pandemic.aspx
References
Davanloo P, Studier FW, et al. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 81:2035-9 (1984). PMID: 6371808
George JT, Srivatsan SG. Posttranscriptional chemical labeling of RNA by using bioorthogonal chemistry. Methods. 120:28-38 (2017). PMID: 28215631
George JT , Srivatsan SG . Bioorthogonal chemistry-based RNA labeling technologies: evolution and current state. Chem Commun (Camb). 56:12307-12318 (2020). PMID: 33026365
Goldberg IH, Rabinowitz M. Comparative utilization of pseudouridine triphosphate and uridine triphosphate by ribonucleic acid polymerase. J Biol Chem. 238:1793-800 (1963). PMID: 13948670
Holstein JM, Rentmeister A. Current covalent modification methods for detecting RNA in fixed and living cells. Methods. 98:18-25 (2016). PMID: 26615954
Huang Y, Eckstein F, et al. Mechanism of ribose 2'-group discrimination by an RNA polymerase. Biochemistry. 1997 36:8231-42. PMID: 9204868
Karikó K, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 23:165-75 (2005). PMID: 16111635
Masaki Y, Ito H, et al. Enzymatic synthesis and reverse transcription of RNAs incorporating 2'-O-carbamoyl uridine triphosphate. Chem Commun (Camb). 52:12889-12892 (2016). PMID: 27738673
Milisavljevič N, Perlíková P, et al. Enzymatic synthesis of base-modified RNA by T7 RNA polymerase. A systematic study and comparison of 5-substituted pyrimidine and 7-substituted 7-deazapurine nucleoside triphosphates as substrates. Org Biomol Chem. 16:5800-5807 (2018). PMID: 30063056
Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51-62. PMID: 2482430
Nance KD, Meier JL. Modifications in an Emergency: The Role of N1-Methylpseudouridine in COVID-19 Vaccines. ACS Cent Sci. 7:748-756 (2021). PMID: 34075344
Pavey JB, Lawrence AJ, et al. Synthesis and transcription studies on 5'-triphosphates derived from 2'-C-branched-uridines: 2'-homouridine-5'-triphosphate is a substrate for T7 RNA polymerase. Org Biomol Chem. 2:869-75 (2004). PMID: 15007416
Siegmund V, Santner T, et al. Screening mutant libraries of T7 RNA polymerase for candidates with increased acceptance of 2'-modified nucleotides. Chem Commun (Camb). 48:9870-2 (2012). PMID: 22932771
Wilgenhof S, Van Nuffel AMT, et al. A phase IB study on intravenous synthetic mRNA electroporated dendritic cell immunotherapy in pretreated advanced melanoma patients. Ann Oncol. 24: 2686-2693 (2013). PMID: 23904461