Almost all naturally occurring types of RNAare post-transcriptionally modified. In general, modifications occur on the nucleobases or ribose, including on the 5'-terminal caps, on messenger RNA (mRNA), as well as on other RNAs. Newly formed eukaryotic RNA transcripts are post-transcriptionally modified. The precursor 5'-guanosine-triphosphate (5'-Gppp) undergoes several modifications before transferred into the cytoplasm. Initially, Eukaryotic mRNAs are transcribed by RNA polymerase II as direct copies of a gene that include several non-coding sequences such as introns.
Nuclear RNA experiences three main events during its life:
(i) Addition of a 5' cap,
(ii) Addition of a 3' cap,
(iii) Removal of unwanted intronic sequences by a process called RNA splicing.
The regulation of the intracellular deoxynucleotide triphosphate (dNTP) pool is essential for genomic stability.
The chemical nature of the 5′ end of RNA appears to determine the stability of RNA during RNA processing, and localization, as well as the translation efficiency. This type of modification provides an additional layer of gene regulation called "epitranscriptomic" gene regulation. A model of a synthetic 7N-Methyl-8-Hydroguanosine-5’-triphosphate capped RNA hexamer is shown in figure 1.
Figure 1: Molecular model of a synthetic 7N-Methyl-8-Hydroguanosine-5'-triphosphate capped RNA hexamer. Images were generated using the structure of vaccinia methyltransferase VP39 complexed with M7G capped RNA hexamer and S-adenosylhomocysteine, PDB 1AV6, and PyMol. The left image shows the cap as van-der-Waals spheres and the nucleotides in the RNA hexamer as sticks. The image to the right illustrates the hexamer as the surface structure.
Recently, Wang et al. developed a system-level method for the quantitative analysis of the cap epitranscriptome called CapQuant. Twenty-one (21) different caps were analyzed. Wang et al. discovered new cap structures in human and mouse cells as well as cell- and tissue-specific variations in cap methylation and high proportions of caps lacking 2'-O-methylation. Also, Wang et al. reported that CapQuant accurately captured the preference for purine nucleotides at eukaryotic transcription start sites as well as the correlation between metabolite levels and metabolite caps.
CapQuant Method
The method for transcriptome-wide quantification of RNA-caps combines off-line HPLC enrichment of cap nucleotides with isotope-dilution, and chromatography-coupled triple-quadrupole mass spectrometry (LC-MS/MS using a TSQ mass spectrometer).
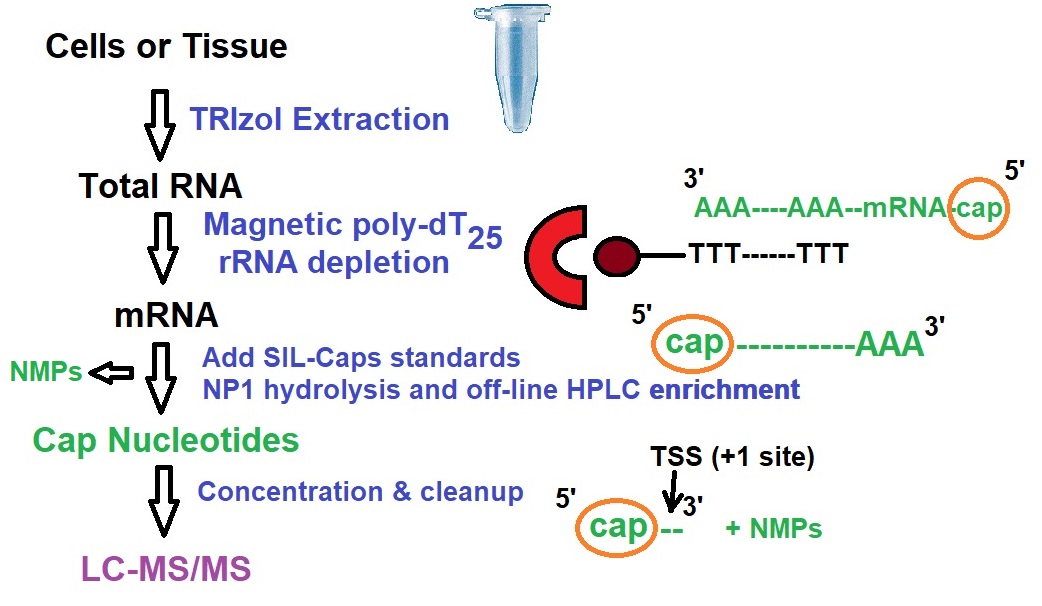
Figure 2: Outline of workflow for CapQuant method for mRNA capture. Legend: SIL, stable isotope label; NP1, Nuclease P1; TSS, Transcription Start Site.
The CapQuant method utilizes nuclease P1 (NP1) for the hydrolysis of RNA to nucleoside monophosphates (NMPs). Di- and tri-phosphate linkages in NpppN and NpppN caps are not hydrolyzed. After NP1 removal, cap structures and 5’-NMPs in the limited digest are analyzed by reversed-phase ion-pairing HPLC and cap-containing fractions are isolated for subsequent quantification using LC–MS/MS.
Reference
Arnold J. Berk; Discovery of RNA splicing and genes in pieces. Proceedings of the National Academy of Sciences Jan 2016, 113 (4) 801-805. [PNAS]
Helm M, Alfonzo JD. Posttranscriptional RNA Modifications: playing metabolic games in a cell's chemical Legoland. Chem Biol. 2014 Feb 20;21(2):174-85. doi: 10.1016/j.chembiol.2013.10.015. Epub 2013 Dec 5. PMID: 24315934; PMCID: PMC3944000. [PMC]
Hodel AE, Gershon PD, Quiocho FA.; Structural basis for sequence-nonspecific recognition of 5'-capped mRNA by a cap-modifying enzyme. Mol Cell. 1998 Feb;1(3):443-7. [PMC]
Wang J, Alvin Chew BL, Lai Y, Dong H, Xu L, Balamkundu S, Cai WM, Cui L, Liu CF, Fu XY, Lin Z, Shi PY, Lu TK, Luo D, Jaffrey SR, Dedon PC. Quantifying the RNA cap epitranscriptome reveals novel caps in cellular and viral RNA. Nucleic Acids Res. 2019 Nov 18;47(20):e130. doi: 10.1093/nar/gkz751. PMID: 31504804; PMCID: PMC6847653. [PMC]