The term “copy number” refers to the number of copies of a particular plasmid per chromosome present in a cell or the number of copies of a gene in the genome of an organism. See medical definition.
Keyaerts et al. in 2004 to 2006 used real-time quantitative RT-PCR for the detection of the SARS-coronavirus. The assay allowed quantitation of virus RNA over a range of 100 to 108 RNA copies per reaction. The use of serial dilutions of standard RNA amplicons established the dynamic range of the assay. A threshold cycle (Ct) value of 37.45 ± 0.04 was reported for 100 copies.
Several technics allow the determination of copy numbers for qPCR samples, for example, for plasmids. These are agarose gel electrophoresis and quantitative polymerase chain reaction (qPCR). Of these two, qPCR is the more sensitive and less time-consuming method. PCR amplicons in the range of 70 to 100 base pairs result in the most efficient amplification.
Anindyajati et al. in 2016 reported a method for copy number determination for plasmids. Using this method, the plasmid copy number can be determined by comparing the quantification signal from the plasmid to the one from the chromosome. The copy number is calculated by using a known copy number plasmid as a standard. Two pairs of primers used in the study, called tdk and ori, were designed for targeting a single gene tdk in the chromosome and a conserved domain in the plasmid’s ori, respectively. The plasmid copy number equals copies per cell.
Ji et al., in 2012, reported a method for the measurement of the gene copy number in biological samples without the need for control samples of known copies.
Determination of threshold cycle
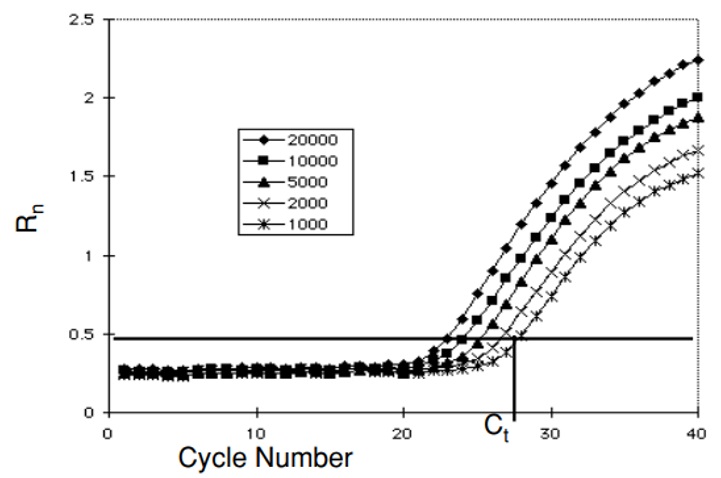
Initial concentration effect on the Rn (normalized reporter signal) value versus cycle number.
Within the exponential phase of PCR at a cycle, the amount of product is proportional to the initial number of template copies.
Threshold Cycle (Ct).
The threshold cycle occurs where the sequence detection application of a PCR instrument begins to detect the increase in signal associated with exponential growth of PCR product.
The Ct is determined by the system by collecting data from the first few PCR cycles and by calculating the average Rn and the standard deviation of the Rn of these cycles. In most PCR systems, the first 15 cycles represent the baseline. The algorithm used by the system multiplies the standard deviation of the background Rn by a default factor of 10 to define the threshold and to search for a point that exceeds the baseline by the value of the threshold. The cycle at which this point occurs is defined as Ct. Ct is dependent on the starting template copy number, and the efficiency of both the DNA amplification the PCR system and the cleavage of the fluorogenic probe used.
Determination of initial concentration of unknowns
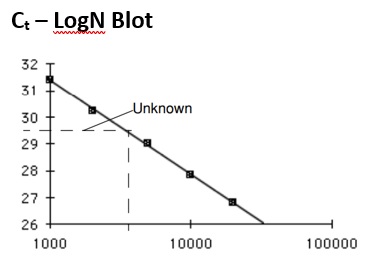
A standard curve of five knowns allows to find the initial concentration of an unknown sample for a given Ct value by interpolation of its initial concentration (see graph).
Since Rn increases as a concentration of amplified product during the exponential growth stage, the relationship of amplified PCR product to the initial template concentration can be expressed as follows:
Nc = N(1 + E)c
where Nc is the concentration of amplified product at any cycle, N is the initial concentration of target template, E is the efficiency of the system, and c is the cycle number.
Example of Data Analysis using a chromosome as a standard
To use a chromosome as a standard primer sets are designed targeting a single copy DNA sequence in either plasmid or chromosome. The cycle threshold (Ct) and the log of the cell number is plotted to determine the ratio of plasmid to chromosome per cell (P/C) based on the following equation.
P/C = (EcCtc / EpCtp)
(Number of plasmid (P))/(Number of chromosome (C) = (EcCtc / EpCtp)
Where, Ec = Ptdk efficiency targeting chromosome. Ptdk, is specific to the thymidine kinase, a single copy gene in E. coli BL21(DE3) chromosome (NCBI, Access No. CP001509.3). Ep = Pori efficiency targeting plasmid. Ctc = Ct of Ptdk targeting chromosome, Ctp = Ct of Pori targeting plasmid.
Adding a plasmid with a known copy number as a standard allows the determination of the unknown copy number.
For an accurate method (often called in-house or LDT method) the following factors need to be taken into account:
(a) Analytical sensitivity (Limit of detection or LOD);
(b) Analytical Specificity for the assay system used;
(c) Results or performance characterisitics. Do they reflect the accurate status of the target studied; and
(d) Extraction method depending on sample type.
Steps needed:
1. Determine quantity of DNA or RNA via fluorometry.
2. Prepare dilution series to determine LOD.
Conversion of Copy number to ngs
For double-stranded DNA (660 g/mole = average mass of 1 bp dsDNA):
Number of copies ( = molecules ) = ( X ng * 6.02214 x 1023 molecules/mole)/ ((N * 660 g/mole) * 1 x 100 ng/g)
Where X = amount of amplicon in nanogram (ng), N = length of double-stranded
DNA amplicon.
For single-stranded RNA (340 g/mole = average mass of 1 nt ssRNA):
Number of copies (#c) = (x ng x 6.0221 x 1023 molecules/mol)/(N x 340 g/mole) )
N = length of single-stranded RNA amplicon.
For a standard with a molecular weight of 100,000 the following number of molecules are found in the specific dilution.
|
|
|
|
Molecules/mol
|
|
|
g/mol
|
100,000.00
|
100
|
6.02214 x 1023
|
S
|
|
mg/mmol
|
100,000.00
|
10-3
|
6.02214 x 1020
|
D1
|
|
μg/μmol
|
100,000.00
|
10-6
|
6.02214 x 1017
|
D2
|
|
ng/nmol
|
100,000.00
|
10-9
|
6.02214 x 1014
|
D3
|
|
pg/pmol
|
100,000.00
|
10-12
|
6.02214 x 1011
|
D4
|
|
fg/fmol
|
100,000.00
|
10-15
|
6.02214 x 108
|
D5
|
|
ag/amol
|
100,000.00
|
10-18
|
6.02214 x 105
|
D6
|
|
zg/zmol
|
100,000.00
|
10-21
|
6.02214 x 102
|
D7 602.214
|
|
|
|
10-23
|
6.02214 x 100
|
D8 ~6 copies/L
|
|
yg/ymol
|
100,000.00
|
10-24
|
6 molecules in 10 L
|
D9
|
According to the FDA, the limit of detection (LOD) is the smallest measured concentration of an analyte from which it is possible to deduce the presence of the analyte in the test sample with acceptable certainty.
Limit of detections for FDA-approved clinical assays are commonly expressed as 50% tissue culture infectious dose per milliliter, or TCID50/ml. This LODs must be converted experimentally to LODs reported as copy numbers per μl for RT-qPCR by preparing dilution series of infectious agents for analysis.
Reference
ABI Prism® 7700 Sequencing Detection System. User Manual.
Anindyajati, Artarini AA, Riani C, Retnoningrum DS. Plasmid Copy Number Determination by Quantitative Polymerase Chain Reaction. Sci Pharm. 2016 Feb 14;84(1):89-101. doi: 10.3797/scipharm.ISP.2015.02. [PMC]
Els Keyaerts, Leen Vijgen, Piet Maes, Griet Duson, Johan Neyts, Marc Van Ranst; Viral load quantitation of SARS-coronavirus RNA using a one-step real-time RT-PCR. International Journal of Infectious Diseases. Volume 10, Issue 1, 2006, 32-37. [PMC]
Gupta, H., Macete, E., Bulo, H., Salvador, C., Warsame, M., Carvalho, E....Mayor, A. (2018). Drug-Resistant Polymorphisms and Copy Numbers in Plasmodium falciparum, Mozambique, 2015. Emerging Infectious Diseases, 24(1), 40-48. [CDC]
Ingram DJ: The study of transgene copy number and organization. Methods in Molecular Biology, vol 286: Transgenic Plants: Methods and Protocols. Edited by L Pena. Totowa, NJ, Humana Press Inc., 2005, pp. 273–289. [PubMed]
Smith M. Validating Real-Time Polymerase Chain Reaction (PCR) Assays. Encyclopedia of Virology. 2021:35–44. [PMC]
Wan Ji, Jian Li, and Jinfeng Liu; A Method for Measuring Gene Copy Number in Biological Samples without Using Control Samples of Known Copies. The Journal of Molecular Diagnostics, Vol. 14, No. 3, May 2012. 280-285. [PubMed]
---...---