With the gradual decline in the number of COVID-19 infections (total of ~103 million cases in the U. S. alone during the 3-year pandemic), the attention is returning to flu, which is projected to incur 25-51 million cases in the U. S. during 2022-2023 (https://www.cdc.gov/flu/about/burden/preliminary-in-season-estimates.htm ). Globally, the influenza virus, which causes flu, infects 2–10% of the entire population, causing 250,000-500,000 deaths annually (El Ramahi et al., 2019). Unlike COVID-19, flu can impact the younger and healthier populations. The disease can deteriorate further by contracting pneumonia caused by the virus or bacterial infection, which may lead to death. Since 1900, the influenza virus has caused pandemics 5 times, with the most notable one being ‘The Spanish flu (1918)’, which may have led to as many as 50 million deaths globally.
For cancer patients, the death rate due to flu is significantly higher than the general public, i.e. 17.4% versus 1.3% (per 100,000 person-years), respectively. ‘Person-year’ is defined as the number of individuals multiplied by the study duration in years (El Ramahi et al., 2019). Over 70% of cancer patients, who contracted flu, received chemotherapy within the preceding 1 month (possibly immunocompromised) during the influenza pandemic in 2009. Those with lung cancer suffered the worst outcome.
There are 4 types of influenza virus: A, B, C, and D. Of these, influenza A and B are capable of infecting humans. To infect, the virus uses its glycoprotein (hemagglutinin) to bind to the sialic acid moiety of saccharides attached to the cell surface. To access the receptor, virally encoded neuraminidase degrades mucus; it may also assist in the release of viruses from a cell. For influenza virus type A, 20 subtypes (H1-H18) of hemagglutinin are known. Neuraminidases are classified into subtypes N1 through N11. Spanish flu was caused by the H1N1 subtype.
Each year, the Center for Disease Control prepares distinct vaccines to prevent seasonal flu in the U. S. The vaccine primarily targets the hemagglutinin antigen of the influenza virus. The need for constantly updating flu vaccine lies in ‘antigenic drift’, which refers to amino acid changes caused by errors introduced during transcription by the virally encoded RNA polymerase (RdRP) (Taubenberger et al., 2010).
Unlike the genome of the coronavirus that caused COVID-19, the genome of the influenza virus consists of 8 segments. The segmented genome allows shuffling of the genomic segments through a process called ‘reassortment’. This could happen if the same cell is infected by more than 1 influenza virus. Through the ‘antigenic shift’, drastically altered antigens can be generated, which could cause a pandemic.
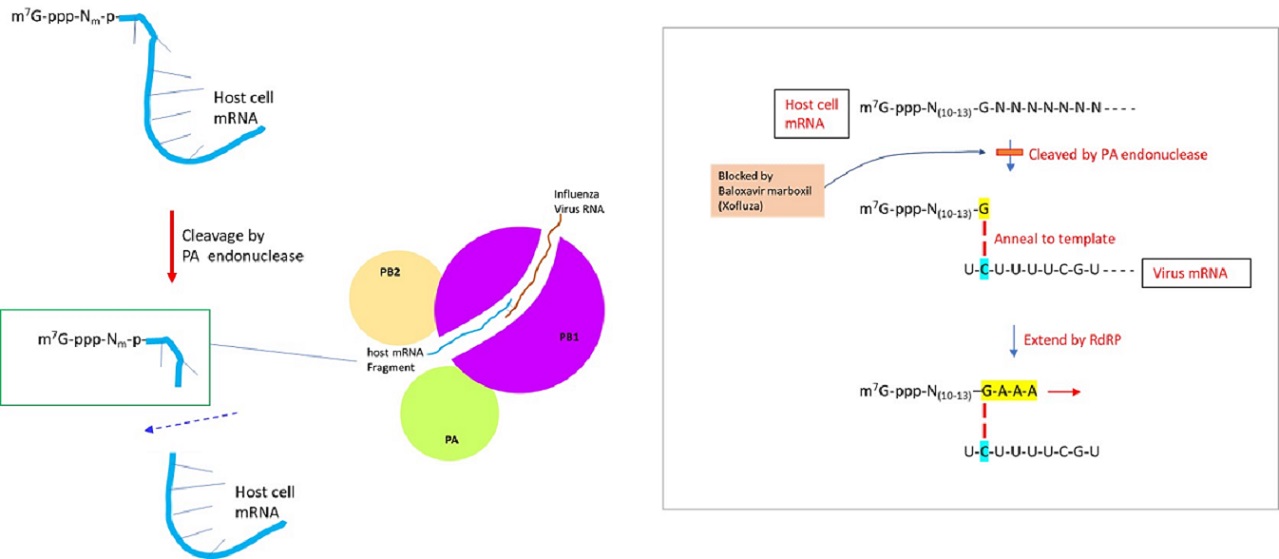
The genome of the influenza virus is composed of negative-sense single-stranded RNA, which is transcribed to generate viral mRNA (to translate into proteins) or cRNA (complementary RNA) to generate vRNA (virus RNA). The positive-sense cRNA template is used to produce the negative-sense genomic RNA of the virus. The synthesis of viral mRNA utilizes the ‘cap-snatching’ mechanism, in which the capped nucleotide primer is derived from the pre-mRNA of the host cell. RNA-dependent RNA polymerase of influenza virus is comprised of 3 subunits: PA, PB1, and PB2. Upon binding of the PB2 subunit to m7G (N7-methyl guanosine)-capped 5’ end of cellular mRNA, the endonuclease activity of the PA subunit cleaves the host pre-mRNA at a position 10-13 nucleotides downstream of the cap structure (De Vlugt et al., 2018). Within the PB1 subunit, the last nucleotide (usually guanidine) of capped host RNA primer is annealed to cytosine of template virus RNA and then extended by RdRP to synthesize mRNA (Li et al., 2020). Recent works indicate that ~55% of the host RNA primers were derived from noncoding RNAs (ex. snRNA U1 and U2) (Li et al., 2020).
Previously, Tamiflu (Gilead Sciences) was developed to inhibit the neuraminidase activity of influenza viruses A and B. More recently, Baloxavir marboxil (Xofluza), a chemical drug that inhibits the cap-dependent endonuclease activity responsible for the cap snatching process was developed. The prodrug is hydrolyzed by arylacetamide deacetylase to release the active agent, baloxavir acid. It was approved by U. S. FDA in 2018 as an antiviral drug to manage the influenza virus. Nevertheless, strains resistant to the drug were detected by 2019 (Yoshino et al., 2019).
The key to preventing an epidemic is the ability to diagnose the infected early to preempt further propagation. For this, Bio-Synthesis, Inc. provides primers and probes (as well as synthetic RNA control) for COVID-19 diagnosis via RT-PCR assay. It specializes in oligonucleotide modification and provides an extensive array of chemically modified nucleoside analogs (over ~200) including bridged nucleic acid (BNA) in addition to mRNA synthesis. A number of options are available to label oligonucleotides (DNA or RNA) with fluorophores either terminally or internally as well as to conjugate to peptides or antibodies. It provides custom conjugation of small molecules such as chemical drugs, metabolites and labeled compounds with synthetic or natural polymers (enzymes, peptide, protein, oligonucleotide, antibody, dendrimer, nanoparticle, etc). It recently acquired a license from BNA Inc. of Osaka, Japan, for the manufacturing and distribution of BNANC, the third generation of BNA oligonucleotides. To meet the demands of therapeutic application, its oligonucleotide products are approaching GMP grade. It has recently entered into collaborative agreement with Bind Therapeutics, Inc. to synthesize miR-21 blocker using BNA for triple-negative breast cancer. The BNA technology provides superior, unequaled advantages in base stacking, binding affinity, aqueous solubility and nuclease resistance. It also improves the formation of duplexes and triplexes by reducing the repulsion between the negatively charged phosphates of the oligonucleotide backbone. Its single-mismatch discriminating power is especially useful for diagnosis (ex. FISH using DNA probe). For clinical application, BNA oligonucleotide exhibits lesser toxicity than other modified nucleotides. For therapeutic consideration, peptide synthesis or modifications may include labeling, conjugation, cyclization, incorporation of unusual amino acids, and modification of side chain and backbone.
https://www.biosyn.com/oligo-flourescent-labeling.aspx
https://www.biosyn.com/tew/Speed-up-Identification-of-COVID19.aspx
https://www.biosyn.com/covid-19.aspx
https://www.biosyn.com/mrna.aspx
https://www.biosyn.com/bioconjugation.aspx
https://www.biosyn.com/tew/Design-Guidelines-for-BNA-based-Oligonucleotide-Probes.aspx#!
Peptide Modifications, Modified Peptide Synthesis - Bio-Synthesis (biosyn.com)
References
De Vlugt C, Sikora D, Pelchat M. Insight into Influenza: A Virus Cap-Snatching. Viruses. 10:641 (2018). PMID: 30453478
El Ramahi R, Freifeld A. Epidemiology, Diagnosis, Treatment, and Prevention of Influenza Infection in Oncology Patients. J Oncol Pract. 15:177-184 (2019). PMID: 30970229
Li L, Dai H, Nguyen AP, Hai R, Gu W. Influenza A virus utilizes noncanonical cap-snatching to diversify its mRNA/ncRNA. RNA. 26:1170-1183 (2020). PMID: 32444459
Taubenberger JK, Kash JC. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe. 7:440-51 (2010). PMID: 20542248
Yoshino R, Yasuo N, Sekijima M. Molecular Dynamics Simulation reveals the mechanism by which the Influenza Cap-dependent Endonuclease acquires resistance against Baloxavir marboxil. Sci Rep. 9:17464 (2019). PMID: 31767949