Heterogeneous nuclear ribonucleoproteins (hnRNPs) isoforms
Heterogeneous nuclear ribonucleoproteins (hnRNPs) comprise a family of RNA-binding proteins. The nature of hnRNPs is very complex and diverse. This multifunctional protein family is involved not only in processing heterogeneous nuclear RNAs (hnRNAs) into mature mRNAs, but also acting as trans-factors in regulating gene expression. Heterogeneous nuclear ribonucleoprotein E1 (hnRNP E1), a subgroup of hnRNPs, is a KH-triple repeat containing the RNA-binding protein. It is encoded by an intronless gene arising from hnRNP E2 through a retrotransposition event. hnRNP E1 is ubiquitously expressed and functions in regulating major steps of gene expression, including pre-mRNA processing, mRNA stability, and translation. This protein family exhibit wide-ranging functions in the nucleus and cytoplasm and interaction with multiple other proteins and appears to be involved in post-transcriptional regulation events or pathways in eukaryotic organisms.
The A/B subfamily of ubiquitously expressed hnRNPs
This gene belongs to the A/B subfamily of ubiquitously expressed heterogeneous nuclear ribonucleo-proteins (hnRNPs). The hnRNPs are RNA binding proteins and complex with heterogeneous nuclear RNA (hnRNA). These proteins are associated with pre-mRNAs in the nucleus and appear to influence pre-mRNA processing and other aspects of mRNA metabolism and transport. While all of the hnRNPs are present in the nucleus, some seem to shuttle between the nucleus and the cytoplasm. The hnRNP proteins have distinct nucleic acid binding properties. The protein encoded by this gene has two repeats of quasi-RRM domains that bind to RNAs. It is one of the most abundant core proteins of hnRNP complexes and it is localized to the nucleoplasm. This protein, along with other hnRNP proteins, is exported from the nucleus, probably bound to mRNA, and is immediately re-imported. Its M9 domain acts as both a nuclear localization and nuclear export signal. The encoded protein is involved in the packaging of pre-mRNA into hnRNP particles, transport of poly A+ mRNA from the nucleus to the cytoplasm, and may modulate splice site selection. It is also thought to have a primary role in the formation of specific myometrial protein species in childbirth. [The myometrium is the middle layer of the uterine wall, mainly consisting of uterine smooth muscle cells.] Multiple alternatively spliced transcript variants have been found for this gene but only two transcripts are fully described. These variants have multiple alternative transcription initiation sites and multiple polyA sites. [From Refseq:
http://www.ncbi.nlm.nih.gov/refseq/].
Precursor mRNA
An immature single strand of messenger ribonucleic acid (mRNA) is called a precursor mRNA (pre-mRNA) which is synthesized in the cell nucleus from a DNA template by transcription. Pre-mRNA makes up the majority of heterogeneous nuclear RNA (hnRNA). hnRNA can also include RNA transcripts that do not end up as cytoplasmic mRNA. More details about these proteins can be found at
http://atlasgeneticsoncology.org/Genes/GC_HNRNPA1.html and
http://www.uniprot.org/uniprot/P09651.
The heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) is sometimes also called “Helix-destabilizing protein”, “Single-strand RNA-binding protein”, or “hnRNP core protein A1” and was identified in the spliceosome C complex, and in a mRNP granule complex, which appears to be at least composed of proteins ACTB, ACTN4, DHX9, ERG, HNRNPA1, HNRNPA2B1, HNRNPAB, HNRNPD, HNRNPL, HNRNPR, HNRNPU, HSPA1, HSPA8, IGF2BP1, ILF2, ILF3, NCBP1, NCL, PABPC1, PABPC4, PABPN1, RPLP0, RPS3, RPS3A, RPS4X, RPS8, RPS9, SYNCRIP, TROVE2, YBX1 and untranslated mRNAs. The protein interacts with SEPT6, with HCV NS5B, and with the 5'-UTR and 3'-UTR of HCV RNA. 5′UTR and 3′UTR are the non-coding regions of HCV RNA, referred herein as 5′ and 3′ untranslated regions that contain important sequence and structural elements critical for HCV translation and RNA replication.
Spliceosome
|
The spliceosome is a multimegadalton RNA-protein machine that removes noncoding sequences from nascent pre-mRNAs. Recruitment of the spliceosome to splice sites and subsequent splicing require a series of dynamic interactions among the spliceosome's component U snRNPs and many additional protein factors.
Pre-mRNA splicing by the U2-type spliceosome
(A) Schematic representation of the two-step mechanism of pre-mRNA splicing. Boxes and solid lines represent the exons (E1, E2) and the intron, respectively. The branch site adenosine is indicated by the letter A and the phosphate groups (p) at the 5′ and 3′ splice sites, which are conserved in the splicing products, are also shown. (B) Conserved sequences found at the 5′ and 3′ splice sites and branch site of U2-type pre-mRNA introns in metazoans and budding yeast (S. cerevisiae). Y = pyrimidine and R = purine. The polypyrimidine tract is indicated by (Yn). (C) Canonical cross-intron assembly and disassembly pathway of the U2-dependent spliceosome. For simplicity, the ordered interactions of the snRNPs (indicated by circles), but not those of non-snRNP proteins, are shown. The various spliceosomal
|
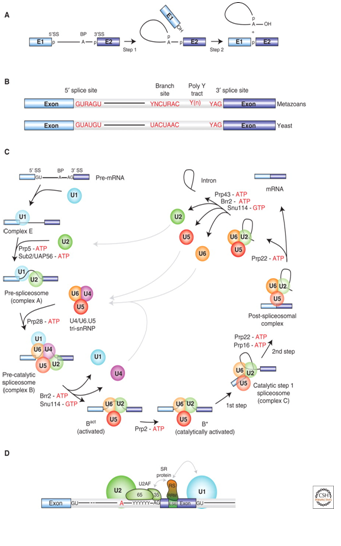 |
complexes are named according to the metazoan nomenclature. Exon and intron sequences are indicated by boxes and lines, respectively. The stages at which the evolutionarily conserved DExH/D-box RNA ATPases/helicases Prp5, Sub2/UAP56, Prp28, Brr2, Prp2, Prp16, Prp22 and Prp43, or the GTPase Snu114, act to facilitate conformational changes are indicated. (D) Model of interactions occurring during exon definition. (From: Will C L , and Lührmann R Cold Spring Harb Perspect Biol 2011;3:a003707).
The DExH/D protein family
The DExH/D protein family is the largest group of enzymes found in eukaryotic RNA metabolism. DExH/D proteins unwind RNA duplexes in an ATP-dependent fashion. In recent years it has become clear that these DExH/D RNA helicases are also involved in the ATP-dependent remodeling of RNA–protein complexes. DExH/D proteins are essential for all aspects of cellular RNA metabolism and processing, for the replication of many viruses and for DNA replication as well. The DExH/D protein family database contains information about these proteins and makes it available over the WWW (
http://www.columbia.edu).
Exon definition
During exon definition exons are recognized and defined as units during early assembly by binding of factors to the 3' end of the intron, followed by a search for a downstream 5' splice site. The presence of both a 3' and a 5' splice site in the correct orientation and within 300 nucleotides of one another will allow that stable exon complexes are formed. Concerted recognition of exons may help explain the 300-nucleotide-length maximum of vertebrate internal exons, the mechanism whereby the splicing machinery ignores cryptic sites within introns, the mechanism whereby exon skipping is normally avoided, and the phenotypes of 5' splice site mutations that inhibit splicing of neighboring introns (Roberson et al. 1990).
Cytoplasmic mRNP granules at a glance
(Source:
Stacy L. Erickson and Jens Lykke-Andersen; Cytoplasmic mRNP granules at a glance Journal of Cell Science 124, 293-297).
Eukaryotic gene expression is controlled by translation and mRNA degradation which are important in the regulation of these processes. Translation and steps in the major pathway of mRNA decay are in competition with each other. mRNAs that are not engaged in translation can aggregate into cytoplasmic mRNP granules referred to as processing bodies (P-bodies) and stress granules, which are related to mRNP particles that control translation in early development and neurons. The analyses of P-bodies and stress granules suggest a dynamic process. This process is referred to as the mRNA Cycle, wherein mRNPs can move between polysomes, P-bodies and stress granules although the functional roles of mRNP assembly into higher order structures remain poorly understood. (Source: Carolyn J. Decker and Roy Parker; P-Bodies and Stress Granules: Possible Roles in the Control of Translation and mRNA Degradation. Cold Spring Harb Perspect Biol a012286 First published online July 3, 2012).
Structure of the Heterogeneous nuclear ribonucleoprotein A1 isoform [Homo sapiens]
(Source: pdb 2LYV)
Chain A, Solution Structure Of The Two Rrm Domains Of Hnrnp A1 (up1) Using Segmental Isotope Labeling
PubMed Abstract: “Human hnRNP A1 is a multi-functional protein involved in many aspects of nucleic-acid processing such as alternative splicing, micro-RNA biogenesis, nucleo-cytoplasmic mRNA transport and telomere biogenesis and maintenance. The N-terminal region of hnRNP A1, also named unwinding protein 1 (UP1), is composed of two closely related RNA recognition motifs (RRM), and is followed by a C-terminal glycine rich region. Although crystal structures of UP1 revealed inter-domain interactions between RRM1 and RRM2 in both the free and bound form of UP1, these interactions have never been established in solution. Moreover, the relative orientation of hnRNP A1 RRMs is different in the free and bound crystal structures of UP1, raising the question of the biological significance of this domain movement. In the present study, we have used NMR spectroscopy in combination with segmental isotope labeling techniques to carefully analyze the inter-RRM contacts present in solution and subsequently determine the structure of UP1 in solution. Our data unambiguously demonstrate that hnRNP A1 RRMs interact in solution, and surprisingly, the relative orientation of the two RRMs observed in solution is different from the one found in the crystal structure of free UP1 and rather resembles the one observed in the nucleic-acid bound form of the protein. This strongly supports the idea that the two RRMs of hnRNP A1 have a single defined relative orientation which is the conformation previously observed in the bound form and now observed in solution using NMR. It is likely that the conformation in the crystal structure of the free form is a less stable form induced by crystal contacts. Importantly, the relative orientation of the RRMs in proteins containing multiple-RRMs strongly influences the RNA binding topologies that are practically accessible to these proteins. Indeed, RRM domains are asymmetric binding platforms contacting single-stranded nucleic acids in a single defined orientation. Therefore, the path of the nucleic acid molecule on the multiple RRM domains is strongly dependent on whether the RRMs are interacting with each other.” (Taken from:
http://www.rcsb.org/pdb/explore/explore.do?structureId=2LYV).
The alignment of 3 hnRNP A1 protein sequences is shown below
Protein and Peptide Library Information for hnRNP proteins
Solid phase
peptide synthesis allows the design and creation of libraries compromising a collection of synthetic peptides. Synthetic libraries have been and can be successfully used for antibody epitope mapping, the determination of the specificity of antibodies, identification of bioactive peptides, development of biological assays, T-cell epitope mapping, vaccine efficacy testing, the screening for ligand-binding activities, screening for antimicrobial peptide activities, peptide-protein interactions, drug discovery, for LC-MS/MS method development and validation as well as for receptor-ligand studies, cellular assays, the study of constrained peptides, modified peptides such as modified histone peptides or the screening for MHC class I and class II peptides, and others.
Furthermore peptide libraries provide synthetic, crude or purified peptides that can be customized for the development of screening applications such as epitope mapping, phosphorylation site identifications or the identification of other post-translational modification (PTM) sites, peptide target interaction studies, mid- to high-throughput selected reaction monitoring (SRM) and multiple reaction monitoring (MRM) assays in quantitative mass spectrometry (MS) workflows.
Peptide libraries for proteomics
The study of proteomes, sub-proteomes and protein pathways often requires quantitative MS analysis that depends on the identification and validation of SRM and MRM assays. Peptide libraries offer great convenience and flexibility in the development of multiple applications involving large numbers of peptides including libraries for quantitative MS approaches. The use of peptide libraries greatly reduces the setup time of MS experiments. The SRMAtlas (http://www.mrmatlas.org/) which attempts to map the entire human proteome can be used as a guide for the development of new types of peptide libraries.
Peptides libraries can be derived from the target protein, and, as an option for the use in MS workflows, with either arginine (R) or lysine (K) as the C-terminal amino acid. Synthetic libraries can cover the most commonly used tryptic proteotypic peptides useful for SRM assay development or proteolytic peptides of the whole protein. The maximum peptide length for screening libraries can range from short peptides (5 amino acids) to 20 amino acids. However, for peptide libraries used in MS workflows the standard maximal length is usually 25 amino acids but this length can be increased up to 35 amino acids to ensure that the vast majority of potential proteotypic peptides that are screened for their suitability for a reliable SRM assay are covered.
Highlights:
- Convenient – peptides are provided in individual tubes or in 96-well plates
- Application-specific – C-terminal amino acid of each peptide can be either R or K or any other amino acid
- Easy to use – peptides can be delivered lyophilized or suspended in 0.1% trifluoroacetic acid (TFA) in 50% (v/v) acetonitrile/water
- Flexible – extensive list of available modifications
Includes:
- Fully synthetic peptides
- Standard mass spectrometric quality control (QC) analysis
- Optional modifications, peptide sizes and levels of QC analysis
- Provided in either in single tubes or 96-well plates
Example of a peptide library for the hnRNP protein useful for MS workflows
Hnrnp A1 protein (up1)
pI of Protein: 8.0,
Protein MW: 22246,
Amino Acid Composition: A10 C2 D13 E18 F10 G17 H8 I8 K16 L8 M4 N4 P6 Q7 R16 S16 T12 V17 W1 Y4
Functional categorization of the proteins for which targeted proteomic assays are available can be found in the MRMAtlas database
|
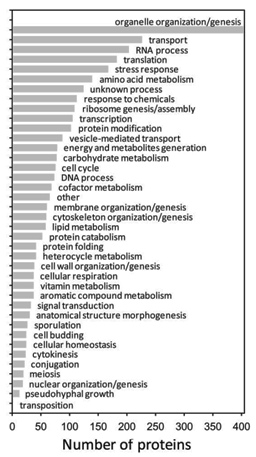
Piccoti et al. in 2008 presented the first database of validated SRM assays for ~1500 yeast proteins. The database was constructed by merging the results of more than 650 SRM-triggered MS2 analyses of S. cerevisiae protein digests, carried out on a triple quadrupole-type mass spectrometer. 1324 proteins are represented by assays for at least one of their peptides proteotypic peptides (PTP’s). The database also contains assays for a small number of peptides common to a maximum of two proteins. The peptides were selected because they show intense signal response by electrospray ionization mass spectrometry. Peptide identifications were validated by collecting a full tandem mass spectrum of the peptides in the QQQ-like mass spectrometer also used for SRM measurements. The database is at present the largest resource of validated SRM/MRM assays of any organism. It currently contains assays for 22% of the yeast proteome and the coverage.
The database contains assays for yeast proteins involved in all biological processes, as defined by gene ontology (GO) nomenclature. Peptides for proteins spanning all ranges of abundance in yeast are present in the dataset, down to a concentration below 50 molecules/cell.
|
 |
The MRMAtlas database
The dataset can be found in the MRMAtlas (
www.mrmatlas.org or
www.srmatlas.org) which is publicly accessible. This database
was created as part of the PeptideAtlas project (
www.peptideatlas.org) and can be queried via the web-interface for peptides,
individual proteins, protein sets, or cellular pathways. These data sets can be used for the design of targeted peptide libraries
that can be custom synthesized. If desired, all or selected peptides can be labeled with stable isotopes useful for spiking
experiments.
Examples of peptide libraries for hnRNP proteins for screening work flows
A: Rrm Domains of HnRNP A1 (up1): Structure ID: PDB: 2LYV_A
Sequence in FASTA format
>gi|433286562|pdb|2LYV|A Chain A, Solution Structure Of The Two Rrm Domains Of Hnrnp A1 (up1) Using Segmental "Isotope Labeling"
GGSKSESPKEPEQLRKLFIGGLSFETTDESLRSHFEQWGTLTDCVVMRDPNTKRSRGFGFVTYATVEEVDAAMNARPHKVD
GRVVEPKRAVSREDSQRPGAHLTVKKIFVGGIKEDTEEHHLRDYFEQYGKIEVIEIMTDRGSGKKRGFAFVTFDDHDSVDK
IVIQKYHTVNGHNCEVRKALSKQEMASASSSQRGR |
Peptide library of 15mers with an overlap of 11 amino acids for HnRNP A1 (up1)
B: Heterogeneous nuclear ribonucleoprotein A1 isoform a [Homo sapiens]
Sequence in FASTA format
>gi|4504445|ref|NP_002127.1| heterogeneous nuclear ribonucleoprotein A1 isoform a [Homo sapiens]
MSKSESPKEPEQLRKLFIGGLSFETTDESLRSHFEQWGTLTDCVVMRDPNTKRSRGFGFVTYATVEEVDAAMNARPHKVD
GRVVEPKRAVSREDSQRPGAHLTVKKIFVGGIKEDTEEHHLRDYFEQYGKIEVIEIMTDRGSGKKRGFAFVTFDDHDSVD
KIVIQKYHTVNGHNCEVRKALSKQEMASASSSQRGRSGSGNFGGGRGGGFGGNDNFGRGGNFSGRGGFGGSRGGGGYGGS
GDGYNGFGNDGSNFGGGGSYNDFGNYNNQSSNFGPMKGGNFGGRSSGPYGGGGQYFAKPRNQGGYGGSSSSSSYGSGRRF |
Peptide library of 15mers with an overlap of 11 amino acids for hnRP A1 a, human
C: Heterogeneous nuclear ribonucleoprotein A1 isoform b [Homo sapiens] {hnRNP A1 b, human}
Sequence in FASTA format
>gi|14043070|ref|NP_112420.1| heterogeneous nuclear ribonucleoprotein A1 isoform b [Homo sapiens]
MSKSESPKEPEQLRKLFIGGLSFETTDESLRSHFEQWGTLTDCVVMRDPNTKRSRGFGFVTYATVEEVDAAMNARPHKVD
GRVVEPKRAVSREDSQRPGAHLTVKKIFVGGIKEDTEEHHLRDYFEQYGKIEVIEIMTDRGSGKKRGFAFVTFDDHDSVD
KIVIQKYHTVNGHNCEVRKALSKQEMASASSSQRGRSGSGNFGGGRGGGFGGNDNFGRGGNFSGRGGFGGSRGGGGYGGS
GDGYNGFGNDGGYGGGGPGYSGGSRGYGSGGQGYGNQGSGYGGSGSYDSYNNGGGGGFGGGSGSNFGGGGSYNDFGNYNN
QSSNFGPMKGGNFGGRSSGPYGGGGQYFAKPRNQGGYGGSSSSSSYGSGRRF |
Peptide library of 15mers with an overlap of 11 amino acids for hnRNP A1 b, human
References
Barraud P, Allain FH;
Solution structure of the two RNA recognition motifs of hnrnp a1 using segmental isotope labeling: how the relative orientation between rrms influences the nucleic acid binding topology. J.Biomol.Nmr (2013) 55 p.119.
Carolyn J.
Decker and Roy Parker; P-Bodies and Stress Granules: Possible Roles in the Control of Translation and mRNA Degradation.
Cold Spring
Harb Perspect Biol a012286 First published online July 3, 2012.
Frank
Desiere, Eric W. Deutsch, Alexey I. Nesvizhskii, Parag Mallick, Nichole King, Jimmy K. Eng, Alan Aderem, Rose Boyle, Erich Brunner, Samuel Donohoe, Nelson Fausto, Ernst Hafen, Lee Hood, Michael G. Katze, Kathleen Kennedy, Floyd Kregenow, Hookeun Lee, Biaoyang Lin, Dan Martin, Jeff Ranish, David J. Rawlings, Lawrence E. Samelson, Yuzuru Shiio, Julian Watts, Bernd Wollscheid, Michael E. Wright, Wei Yan, Lihong Yang, Eugene Yi, Hui Zhang and Ruedi Aebersold Genome Biology 2004, 6:R9
Integration of Peptide Sequences Obtained by High-Throughput Mass Spectrometry with the Human Genome.
Eric W
Deutsch, Henry Lam & Ruedi Aebersold EMBO reports 9, 5, 429–434 (2008)
PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows
Domon B, Aebersold R. Science. 2006 Apr 14;312(5771):212-7
Mass spectrometry and protein analysis.
Lange V,
Malmström JA, Didion J, King NL, Johansson BP, Schäfer J, Rameseder J, Wong CH, Deutsch EW, Brusniak MY, Bühlmann P, Björck L, Domon B, Aebersold R. Mol Cell Proteomics. 2008 Apr 13.
Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring.
Paola
Picotti, Mathieu Clément-Ziza, Henry Lam, David S. Campbell, Alexander Schmidt, Eric W. Deutsch, Hannes Röst, Zhi Sun, Oliver Rinner, Lukas Reiter, Qin Shen, Jacob J. Michaelson, Andreas Frei, Simon Alberti, Ulrike Kusebauch, Bernd Wollscheid, Robert L. Moritz, Andreas Beyer & Ruedi Aebersold
Nature. 2013 Feb 14;494(7436):266-70. doi: 10.1038/nature11835. Epub 2013 Jan 20.
A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis
Paola
Picotti, Henry Lam, David Campbell, Eric W. Deutsch, Hamid Mirzaei, Jeff Ranish, Bruno Domon and Ruedi Aebersold Nature Methods
A database of mass spectrometric assays for the yeast proteome. Nat Methods. 2008 November; 5(11): 913–914.
B L
Robberson, G J Cote, and S M Berget; Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 January; 10(1): 84–94. PMCID: PMC360715
Other resources
HNRNPA1: provided by
HGNC (http://www.genenames.org/), heterogeneous nuclear ribonucleoprotein A1: provided by
HGNC. Primary source:
HGNC:5031, See related:
Ensembl:ENSG00000135486;
HPRD:01242;
MIM:164017;
Vega:OTTHUMG00000169702; Gene type: protein coding; Organism:
Homo sapiens. Lineage: Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi; Mammalia; Eutheria; Euarchontoglires; Primates; Haplorrhini; Catarrhini; Hominidae; Homo. Also known as: HNRPA1; HNRPA1L3; hnRNP A1; hnRNP-A1