Peptide-based therapies offer a cost-effective and rapidly developed therapeutic response to pandemic outbreaks such as COVID-19. However, one drawback is their proteolytic instability. Peptidomimetics, the art of designing molecules that mimic natural peptides, promises to overcome these hurdles.
For the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2; COVID-19), therapeutic peptides and peptide-based therapeutics may target main viral entry pathways into human cells. One involves the interaction between the host angiotensin-converting enzyme-2 (ACE2) receptor protein and the viral spike or surface (S) protein.
The S protein allows the coronavirus to bind to the host cell receptor ACE2. Peptides designed to interfere with the binding event can inhibit viral entry, thereby preventing infection. These type of peptides are also known as antiviral-peptides. Antiviral peptides inhibit the binding of the SARS coronavirus spike RBD to the cellular receptor, ACE2. These peptides are also called "Coronavirus Inhibitory Peptides." However, to stabilize functional natural peptides peptide mimetics will need to be designed. For example, stapled peptides allow the design and synthesis of conformationally constrained peptides.
Table 1: SARS-CoV-2 (COVID-19) Peptides
|
Peptide
|
Target
|
Sequence: SARS-CoV-2
|
|
hACE2 n-terminal peptide
AA30 to 53. SBP1 (peptidase domain of ACE2)
|
SARS-CoV-2 S protein. S protein-RBD
|
IEEQAKTFLDKFNHEAEDLFYQSS
|
|
SARS-CoV-2 S RBD
|
ACE2 receptor
|
rvqptesivrfpnitnl
|
|
SARS-CoV-2 peptide AA483 to 506
|
ACE2 receptor
|
VEGFNCYFPLQSYGFQPTNGVGYQ
|
|
2019-nCoV-HR2P
|
HR1 domain of CoV S protein
|
DISGINASVVNIQKEIDRLNEVAKNLNESLIDLQEL
|
|
EK1
|
HR1 domain of CoV S protein
|
SLDQINVTFLDLEYEMKLEEAIKLEESYIDLKEL
|
|
EK1C4 (lipopeptide)
|
HR1 domain of CoV S protein
|
(N)EK1-GSGSG-PEG4-(Chol)
|
|
IPB02
|
HR1 domain of CoV S protein
|
ISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELK (Cholesterol)
|
{ COVID-19 glycopeptides: Xia, S., Liu, M., Wang, C. et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res 30, 343–355 (2020). [Nature] }
The following models of the coronavirus SARS-CoV-2 structure and mechanism of entry allow the selection of target molecules useful for diagnostics and vaccine development.
Coronavirus cell entry mechanisms
|
Virus
|
Primary Entry Event
|
Secondary Entry Event
|
|
SARS-CoV
|
Binding to host peptidases as their entry receptors.
Target: angiotensin-converting enzyme-2 (ACE2).
|
Possibly S protein mediated internalization through C-type lectins recognizing high-mannose glycans on the S protein via endocytosis mechanism and viral membrane fusion occurring in endosomes.
|
|
MERS-CoV
|
Binding to host peptidases as their entry receptors.
Target: dipeptidyl peptidase-4 (DPP4)
|
Possibly S protein mediated internalization through C-type lectins recognizing high-mannose glycans on the S protein via endocytosis mechanism and viral membrane fusion occurring in endosomes.
|
|
SARS-CoV-2
|
Binding to host peptidases as their entry receptors. Target: angiotensin-converting enzyme-2 (ACE2)
|
Possibly S protein mediated internalization through C-type lectins recognizing high-mannose glycans on the S protein via endocytosis mechanism and viral membrane fusion occurring in endosomes.
|
|
Other coronaviruses
|
Can recognize post-translational modifications such as glycans or adhesion molecules.
|
|
{ VanPatten S, He M, Altiti A, F Cheng K, Ghanem MH, Al-Abed Y. Evidence supporting the use of peptides and peptidomimetics as potential SARS-CoV-2 (COVID-19) therapeutics. Future Med Chem. 2020 Sep;12(18):1647-1656. doi: 10.4155/fmc-2020-0180. Epub 2020 Jul 16. PMID: 32672061; PMCID: PMC7364852. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7364852/ }
In cell culture, coronaviruses (CoVs) can enter cells via two distinct routes:
(1) the late endosome where the S protein is cleaved by cathepsins, or
(2) via the cell-surface or early endosome utilizing trypsin-like proteases for S cleavage.
Clinical isolates of human 420 CoV (HCoV) achieve activation by trypsin-like serine proteases and utilize endosomal cathepsins only in the absence of suitable trypsin-like proteases in cell culture.
Models of the life cycle for SARS-CoV are thought to be similar for that of SARS-CoV-2 (COVID-19). Both, coronavirus SARS-CoV and SARS-CoV-2 use the angiontensin-converting enzyme 2 (ACE2) as the host receptor. Asian males appear to express more ACE2 in the lungs than Asian women.
(1) Cell entry
To enable cell entry, the virus envelope spike glycoprotein S interacts with the hosts cellular receptor ACE2. Utilizing the endocytosis mechanism of the cell, the virus can enter the cell. In contrast to SARS-CoV, SARS-CoV-2 appears also to enter the cell via fusion with the cell plasma membrane directly.
(2) Uncoating
In the endosome the pH ranging from pH 4.5 to 6.5 as compared to the neutral to slightly basic pH of 7.0 to 7.4 in the cytosol promotes fusion of the endosome membrane with the virus envelope, releasing the nucleocapsid into the cytoplasm. The capsid is degraded by cellular proteases leaving the viral genome as free RNA in the cytoplasm.
{ The endosomal-lysosome system: Hu YB, Dammer EB, Ren RJ, Wang G. The endosomal-lysosomal system: from acidification and cargo sorting to neurodegeneration. Transl Neurodegener. 2015 Sep 30;4:18. doi: 10.1186/s40035-015-0041-1. [PMC] ; Dechant R, Peter M. Cytosolic pH: A conserved regulator of cell growth? Mol Cell Oncol. 2014 Dec 31;1(4):e969643. [PMC] }
(3) Translation
The positive-strand viral RNA is directly translated by the cellular translation machinery, the ribosome. The polyproteins are translated forming the replication and transcription complex after processing.
(4) Synthesis
Synthesis of negative sense pre-genomic RNA: The complementary strand of negative-sense pre-genomic RNA is synthesized which now serves as a template for the replication of the positive-sense viral genome.
(5) Replication
The replication process occurs in the cytoplasm of the cell. Replication and transcription complex synthesize smaller, positive-sense subgenomic RNAs.
(6) Translation
Subgenomic RNAs are translated into viral proteins. Structural viral proteins are synthesized and assembled in the endoplasmic reticulum. The viral envelope originates from the endoplasmic reticulum membrane.
(7) Transport to the cell surface
The assembled viral particle travels through the cellular vesicle transport system including the Golgi apparatus to the cell surface.
(8) Cell exit or budding
Viral particles leave the cell via exocytosis. Viral particles maturate with the help of viral proteases to fit all viral components together and the viral particle is no infectious again. A new infection cycle can begin.
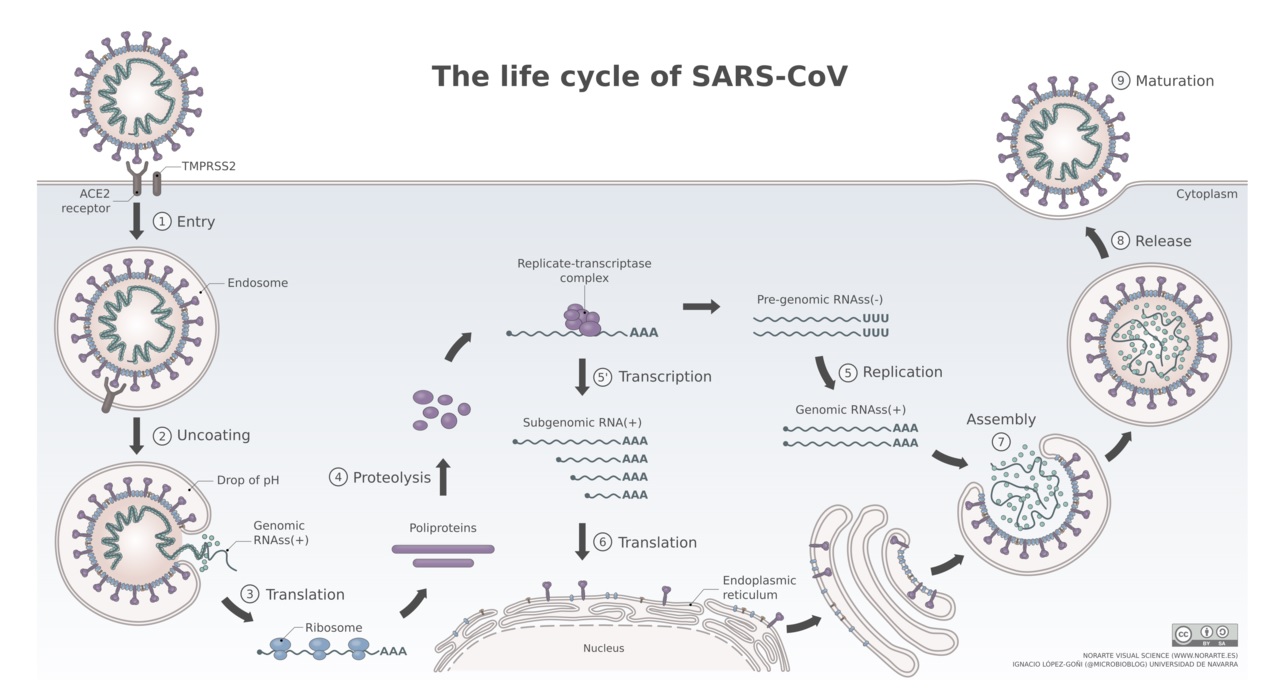
{ Source: By Vega Asensio Wikimedia commons }
The autophagy-related (Atg) gene-encoded protein sytem supports virus replication and viral antigen presentation in major histocompatibility (MHC) class I and II molecules. This sytem may offer a target for the development of antiviral therapies as well.
{ Münz C. Autophagy Proteins in Viral Exocytosis and Anti-Viral Immune Responses. Viruses. 2017 Oct 4;9(10):288. doi: 10.3390/v9100288. [PMC]; de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016 Aug;14(8):523-34. [PMC] }
Early models of viral replications were proposed by Sawicki 2008, den Boon et al. 1996, and van Dinten et al.1997.
{ Sawicki SG. Coronavirus Genome Replication. Viral Genome Replication. 2008 Nov 1:25–39. [PMC] ; den Boon JA, Kleijnen MF, Spaan WJ, Snijder EJ. Equine arteritis virus subgenomic mRNA synthesis: analysis of leader-body junctions and replicative-form RNAs. J Virol. 1996 Jul;70(7):4291-8. [PMC] ; van Dinten LC, den Boon JA, Wassenaar AL, Spaan WJ, Snijder EJ. An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc Natl Acad Sci U S A. 1997 Feb 4;94(3):991-6. [PMC] }
To determine the RNA sequences, direct sequencing of RNA templates was performed using the method from Fichot and Girard.
{ Fichot O, Girard M. An improved method for sequencing of RNA templates. Nucleic Acids Res. 1990 Oct 25;18(20):6162. [PMC] }
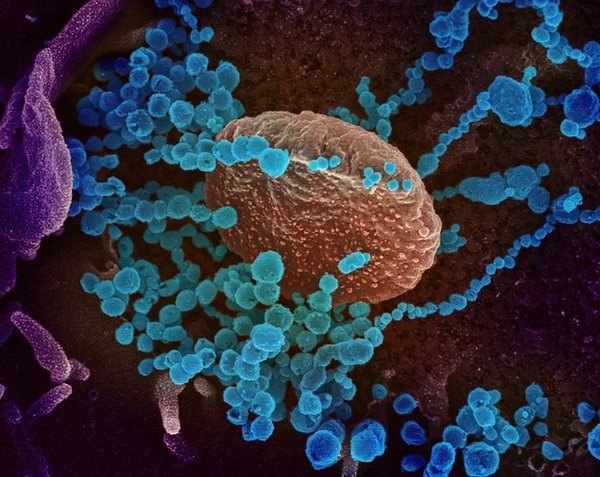
Novel Coronavirus SARS-CoV-2: This image shows SARS-CoV-2 (round blue objects) emerging from the surface of cells cultured in the lab. A scanning electron microscope was used for its generation. The corona virus SARS-CoV-2, or 2019-nCoV, is the cause of COVID-19. The virus shown was isolated from a patient in the U.S. The image was captured and colorized at NIAID's Rocky Mountain Laboratories (RML) in Hamilton, Montana.
{ Credit: NIAID-RML) TEMI Ref: Prasad S, Potdar V, Cherian S, Abraham P, Basu A; ICMR-NIV NIC Team. Transmission electron microscopy imaging of SARS-CoV-2. Indian J Med Res. 2020 Feb & Mar;151(2 & 3):241-243. doi: 10.4103/ijmr.IJMR_577_20 [PMC]. }
Coronavirus SARS-CoV-2 viroid and genome
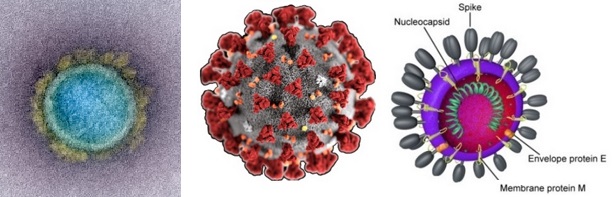
Virion: Enveloped, spherical, 60 to 140 nm in diameter with 9 to 12 nm spikes
Genome: ~ 30 kb positive-sense, ssRNA
RNA Transcript: 5'-cap, 3'-poly-A tail
Proteome: ~ 10 proteins
Transmission: Links to seafood and animal market cases suggest animal-to-human transmission.
Sustained human-to-human transmission observed in later cases.
Phylogeny: Closely related to bat-SL-CoVZC45 and bat-SL-CoVZX21.
The typical organization of the viral genome is
5’-leader-UTR-replicase-S(Spike)-E(Envelope)-M(Membrane)-N(Nucleocapsid)-3’-UTR-poly(A) tail.
Accessory genes are interspersed within the structural genes at the 3’-end of the genome.
Structural models of SARS-CoV-2 S protein
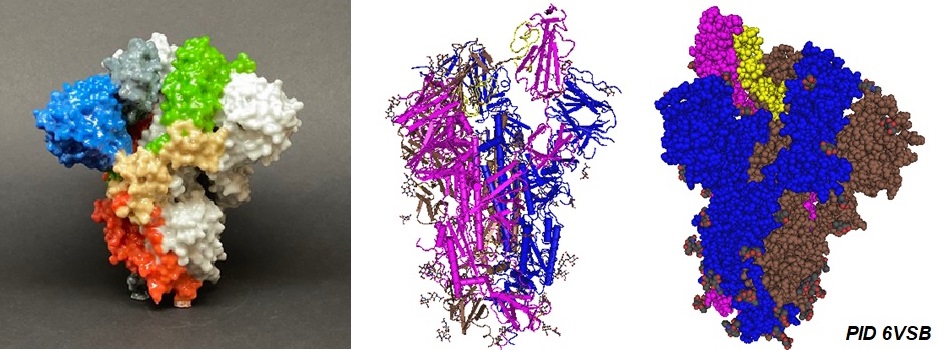
Source: NIAID-RML; Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020 Mar 13;367(6483):1260-1263. [PMC].
ACE2-SARS-CoV-2 interphasing peptide
The angiotensin-converting enzyme 2 (ACE2) is the cellular receptor for the SARS coronavirus (SARS-Cov or SARS-CoV-1) and SARS-CoV-2 S protein. The following peptide is located at the interphase between the ACE2 receptor protein and the receptor binding domain (RBD) of the SARS-CoV-2 S protein. This peptide specifically binds to the SARS-CoV-2 receptor binding domain however with low nano-molar efficiency.
hACE2 n-terminal peptide AA30 to 53:IEEQAKTFLDKFNHEAEDLFYQSS
Figure 1: Structural model of the SARS-Cov-2 RBD-ACE2 complex [PID 6LZG]. The location of the inter-phasing peptide is indicated in yellow. The crystal structure of the C-terminal domain (RBD) of SARS-CoV-2 (SARS-CoV-2-CTD or SARS-Cov-2 RBD) as part of the spike (S) protein in complex with human ACE2 (hACE2) was solved by Wang et al. in 2020. The structural model reveals how human angiotensin hACE2 binds and interacts with the S protein in a mode similar to that observed for SARS-CoV.
{ Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, Wang Q, Zhou H, Yan J, Qi J. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020 May 14;181(4):894-904.e9. [PMC]. / G. Zhang, S. Pomplun, A. R. Loftis, A. Loas, B. L. Pentelute; The first-in-class peptide binder to the SARS-CoV-2 spike protein. bioRxiv 2020.03.19.999318; [bioRχiv] }
SARS-CoV-2 S protein RBD Peptide
Spike protein receptor binding domain, AA319-335 S protein sequence. Protein ID 6XR8.
RVQPTESIVRFPNITNL
Computational approaches allowed the identification of B- and T-cell epitopes for the spike (S) glycoprotein of SARS-CoV-2 by its interactions with the human leukocyte antigen alleles. The study identified 24 peptide stretches on the SARS-CoV-2 S protein well conserved among the reported strains. The structural model of the S protein allows observation of the location of predicted surface peptides. Twenty surface-exposed peptides have reasonable epitope binding efficiency. These identified T-cell epitopes could be candidates for vaccine designs.
{ Vashi Y, Jagrit V, Kumar S. Understanding the B and T cell epitopes of spike protein of severe acute respiratory syndrome coronavirus-2: A computational way to predict the immunogens. Infect Genet Evol. 2020 Oct;84:104382. [PMC] }
SARS-CoV-2 S protein RBD interphasing peptide
This S protein RBD interphase peptide is in contact with the ACE2 receptor protein.
SARS-CoV-2 peptide AA483 to 506: VEGFNCYFPLQSYGFQPTNGVGYQ
.jpg)
Figure 2: CryoEM structure of the SARS-CoV-2 Spike protein in complex with ACE2 (PID 7A96): (Left image) The location of the ACE2 peptide IEEQAKTFLDKFNHEAEDLFYQSS on the interphase with the S protein in the complex is shown in yellow. (Right image) The S protein RBD peptide VEGFNCYFPLQSYGFQPTNGVGYQ in contact with ACE2 in the complex is shown in yellow. The structure as solved by Benton et al. revealed a refolding event of the S1 subdomain after binding to ACE2. The refolding disrupts interactions with the S2 subdomain which destabilized the structure of S2 proximal to the secondary (S2') cleavage site.
{ Benton DJ, Wrobel AG, Xu P, Roustan C, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020 Sep 17. doi: 10.1038/s41586-020-2772-0. Epub ahead of print. PMID: 32942285. [PMC]. }
---...---